3E25
 
 | | Crystal structure of M. tuberculosis glucosyl-3-phosphoglycerate synthase | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, MAGNESIUM ION, Putative uncharacterized protein, ... | | Authors: | Pereira, P.J.B, Empadinhas, N, Costa, M.S, Macedo-Ribeiro, S. | | Deposit date: | 2008-08-05 | | Release date: | 2008-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mycobacterium tuberculosis glucosyl-3-phosphoglycerate synthase: structure of a key enzyme in methylglucose lipopolysaccharide biosynthesis
Plos One, 3, 2008
|
|
3PQR
 
 | | Crystal structure of Metarhodopsin II in complex with a C-terminal peptide derived from the Galpha subunit of transducin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-1, ... | | Authors: | Choe, H.-W, Kim, Y.J, Park, J.H, Morizumi, T, Pai, E.F, Krauss, N, Hofmann, K.P, Scheerer, P, Ernst, O.P. | | Deposit date: | 2010-11-26 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of metarhodopsin II.
Nature, 471, 2011
|
|
7T1T
 
 | | JAK2 JH2 IN COMPLEX WITH JAK292 | | Descriptor: | (2S)-2-[({4-[(2-amino-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy]phenyl}carbamoyl)amino]-4-phenylbutanoic acid, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Ippolito, J.A, Henry, S, Krimmer, S.G, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2021-12-02 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Conversion of a False Virtual Screen Hit into Selective JAK2 JH2 Domain Binders Using Convergent Design Strategies
Acs Med.Chem.Lett., 13, 2022
|
|
7T2X
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S in Complex with 2-chloro-4-((4-hydroxybenzyl)amino)-5-phenylthieno[2,3-d]pyrimidin-6-ol and GRIP Peptide | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, S-(2-chloro-6-{[(4-hydroxyphenyl)methyl]amino}pyrimidin-4-yl) phenylethanethioate | | Authors: | Joiner, C, Sammeta, V.K.R, Norris, J.D, McDonnell, D.P, Wilson, T.M, Fanning, S.W. | | Deposit date: | 2021-12-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A New Chemotype of Chemically Tractable Nonsteroidal Estrogens Based on a Thieno[2,3- d ]pyrimidine Core.
Acs Med.Chem.Lett., 13, 2022
|
|
3DBI
 
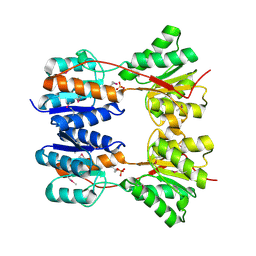 | | CRYSTAL STRUCTURE OF SUGAR-BINDING TRANSCRIPTIONAL REGULATOR (LACI FAMILY) FROM ESCHERICHIA COLI COMPLEXED WITH PHOSPHATE | | Descriptor: | GLYCEROL, PHOSPHATE ION, SUGAR-BINDING TRANSCRIPTIONAL REGULATOR, ... | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Wu, B, Maletic, M, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-01 | | Release date: | 2008-07-01 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Sugar-Binding Transcriptional Regulator (LacI Family) from Escherichia Coli Complexed with Phosphate.
To be Published
|
|
3G04
 
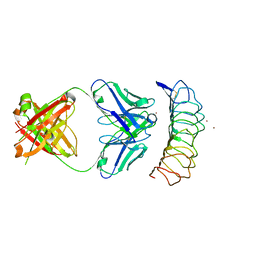 | | Crystal structure of the TSH receptor in complex with a thyroid-stimulating autoantibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HUMAN THYROID STIMULATING AUTOANTIBODY M22 HEAVY CHAIN, HUMAN THYROID STIMULATING AUTOANTIBODY M22 LIGHT CHAIN, ... | | Authors: | Sanders, J, Chirgadze, D.Y, Sanders, P, Baker, S, Sullivan, A, Bhardwaja, A, Bolton, J, Reeve, M, Nakatake, N, Evans, M, Richards, T, Powell, M, Miguel, R.N, Blundell, T.L, Furmaniak, J, Smith, B.R. | | Deposit date: | 2009-01-27 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the TSH receptor in complex with a thyroid-stimulating autoantibody
Thyroid, 17, 2007
|
|
3G33
 
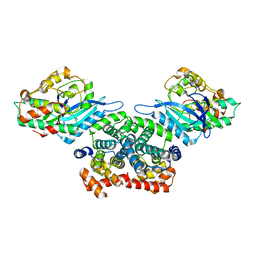 | | Crystal structure of CDK4/cyclin D3 | | Descriptor: | CCND3 protein, Cell division protein kinase 4 | | Authors: | Takaki, T, Echalier, A, Brown, N.R, Hunt, T, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2009-02-01 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of CDK4/cyclin D3 has implications for models of CDK activation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7TJP
 
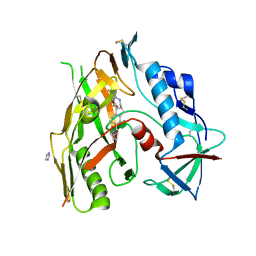 | | HIV-1 gp120 complex with CJF-II-195 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, IMIDAZOLE, ... | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural and Functional Characterization of Indane-Core CD4-Mimetic Compounds Substituted with Heterocyclic Amines
Acs Medicinal Chemistry Letters, 14, 2023
|
|
7TJO
 
 | | HIV-1 gp120 complex with CJF-II-197-S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, IMIDAZOLE, ... | | Authors: | Gong, Z, Hendrickson, W.A. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structural and Functional Characterization of Indane-Core CD4-Mimetic Compounds Substituted with Heterocyclic Amines
ACS Medicinal Chemistry Letters, 14, 2023
|
|
3DS6
 
 | | P38 complex with a phthalazine inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-4-methyl-3-[1-(2-methylphenyl)phthalazin-6-yl]benzamide | | Authors: | Herberich, B, Syed, R, Li, V, Grosfeld, D. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of highly selective and potent p38 inhibitors based on a phthalazine scaffold.
J.Med.Chem., 51, 2008
|
|
7S1N
 
 | |
3ESS
 
 | |
3QQY
 
 | |
7UP6
 
 | | Crystal structure of C-terminal domain of MSK1 in complex with in covalently bound literature RSK2 inhibitor pyrrolopyrimidine cyanoacrylamide compound 25 (co-crystal) | | Descriptor: | (E)-3-(3-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)phenyl)-2-cyanoacrylamide bound form, OXAMIC ACID, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Abendroth, J, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
7UP4
 
 | | Crystal structure of C-terminal Domain of MSK1 in complex with covalently bound pyrrolopyrimidine compound 20 (co-crystal) | | Descriptor: | (5M)-5-(2,5-dichloropyrimidin-4-yl)-5H-pyrrolo[3,2-d]pyrimidine, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Abendroth, J, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
7UP8
 
 | |
7UP5
 
 | | Crystal structure of C-terminal Domain of MSK1 in complex with covalently bound pyrrolopyrimidine compound 23 (co-crystal) | | Descriptor: | (2M)-6-chloro-2-(5H-pyrrolo[3,2-d]pyrimidin-5-yl)pyridine-3-carbonitrile, IODIDE ION, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Edwards, T.E, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
7UP7
 
 | | Crystal structure of C-terminal Domain of MSK1 in complex with covalently bound with literature RSK2 inhibitor indazole cyanoacrylamide compound 26 (soak) | | Descriptor: | (2S)-2-cyano-N-(1-hydroxy-2-methylpropan-2-yl)-3-[3-(3,4,5-trimethoxyphenyl)-1H-indazol-5-yl]propanamide, Ribosomal protein S6 kinase alpha-5 | | Authors: | Yano, J.K, Abendroth, J, Hall, A. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Characterization of a Novel Series of Chloropyrimidines as Covalent Inhibitors of the Kinase MSK1.
Acs Med.Chem.Lett., 13, 2022
|
|
3FEW
 
 | | Structure and Function of Colicin S4, a colicin with a duplicated receptor binding domain | | Descriptor: | Colicin S4, SODIUM ION | | Authors: | Arnold, T, Linke, D, Zeth, K. | | Deposit date: | 2008-12-01 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and Function of Colicin S4, a Colicin with a Duplicated Receptor-binding Domain
J.Biol.Chem., 284, 2009
|
|
3H7V
 
 | | CRYSTAL STRUCTURE OF O-SUCCINYLBENZOATE SYNTHASE FROM THERMOSYNECHOCOCCUS ELONGATUS BP-1 complexed with MG in the active site | | Descriptor: | MAGNESIUM ION, O-SUCCINYLBENZOATE SYNTHASE | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-12 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7PFS
 
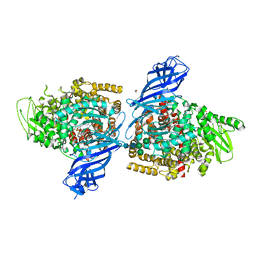 | | Crystal structure of ERAP2 aminopeptidase in complex with phosphinic pseudotripeptide ((1R)-1-Amino-3-phenylpropyl){2-([1,1:3,1-terphenyl]-5-ylmethyl)-3-[((2S)-1-amino-1-oxo-3-phenylpropan-2-yl)-amino]-3-oxopropyl}phosphinic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Stratikos, E, Mpakali, A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibitor-Dependent Usage of the S1' Specificity Pocket of ER Aminopeptidase 2.
Acs Med.Chem.Lett., 13, 2022
|
|
7YX6
 
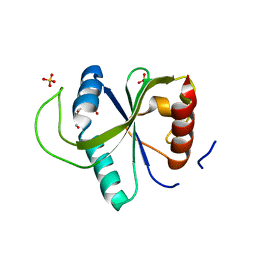 | | Crystal structure of YTHDF2 with compound YLI_DF_027 | | Descriptor: | 6-ethyl-1H-pyrimidine-2,4-dione, GLYCEROL, SULFATE ION, ... | | Authors: | Nai, F, Li, Y, Caflisch, A. | | Deposit date: | 2022-02-15 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
7YXE
 
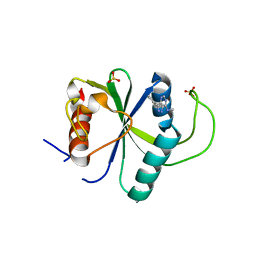 | | Crystal structure of YTHDF2 with compound ZA_143 | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Nai, F, Zalesak, F, Li, Y, Caflisch, A. | | Deposit date: | 2022-02-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
7Z26
 
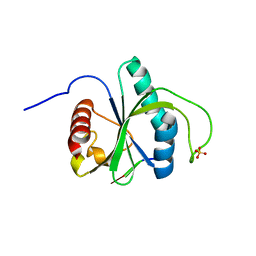 | | Crystal structure of YTHDF2 YTH domain in complex with m6A RNA | | Descriptor: | GLYCEROL, RNA (5'-R(P*(6MZ)P*CP*U)-3'), SULFATE ION, ... | | Authors: | Nai, F, Nachawati, R, Li, Y, Caflisch, A. | | Deposit date: | 2022-02-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
7Z4U
 
 | | Crystal structure of YTHDF2 with compound YLI_DF_028 | | Descriptor: | 6-cyclopropyl-3-methyl-1H-pyrimidine-2,4-dione, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nai, F, Li, Y, Caflisch, A. | | Deposit date: | 2022-03-04 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
