6ICZ
 
 | | Cryo-EM structure of a human post-catalytic spliceosome (P complex) at 3.0 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6ID1
 
 | | Cryo-EM structure of a human intron lariat spliceosome after Prp43 loaded (ILS2 complex) at 2.9 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6IEG
 
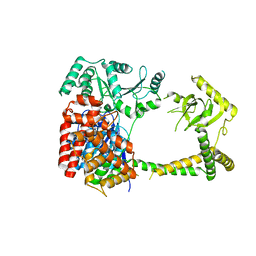 | | Crystal structure of human MTR4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Exosome RNA helicase MTR4, MAGNESIUM ION | | Authors: | Chen, J.Y, Yun, C.H. | | Deposit date: | 2018-09-14 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction.
Genes Dev., 33, 2019
|
|
6IEH
 
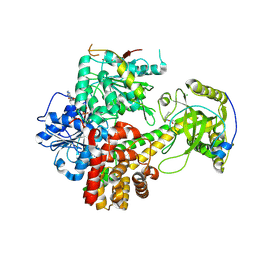 | | Crystal structures of the hMTR4-NRDE2 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Exosome RNA helicase MTR4, ... | | Authors: | Chen, J.Y, Yun, C.H. | | Deposit date: | 2018-09-14 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction.
Genes Dev., 33, 2019
|
|
6KYV
 
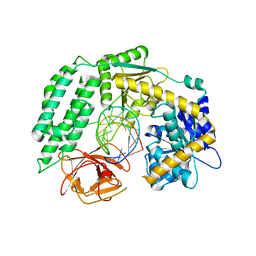 | | Crystal Structure of RIG-I and hairpin RNA with G-U wobble base pairs | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*GP*GP*UP*AP*GP*AP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*UP*UP*GP*CP*C)-3'), ZINC ION | | Authors: | Kim, K.-H, Hwang, J, Kim, J.H, Son, K.-P, Jang, Y, Kim, M, Kang, S.-J, Lee, J.-O, Choi, B.-S. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biophysical properties of RIG-I bound to dsRNA with G-U wobble base pairs.
Rna Biol., 17, 2020
|
|
6L5L
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at apo state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5M
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Nucleolar RNA helicase 2 | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5N
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-unwound state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6L5O
 
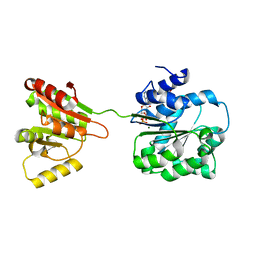 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
6LQS
 
 | |
6M6A
 
 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6M6B
 
 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase and ATP-gamma-S | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
6O16
 
 | | Crystal structure of murine DHX37 in complex with RNA | | Descriptor: | DEAH (Asp-Glu-Ala-His) box polypeptide 37, RNA (5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Boneberg, F, Brandmann, T, Kobel, L, van den Heuvel, J, Bargsten, K, Bammert, L, Kutay, U, Jinek, M. | | Deposit date: | 2019-02-18 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.875 Å) | | Cite: | Molecular mechanism of the RNA helicase DHX37 and its activation by UTP14A in ribosome biogenesis.
Rna, 25, 2019
|
|
6O5F
 
 | | Crystal structure of DEAD-box RNA helicase DDX3X at pre-unwound state | | Descriptor: | ATP-dependent RNA helicase DDX3X, CHLORIDE ION, RNA (5'-R(P*CP*AP*AP*GP*GP*UP*CP*AP*UP*UP*CP*GP*CP*AP*AP*GP*AP*GP*UP*GP*GP*CP*C)-3') | | Authors: | Song, H, Ji, X. | | Deposit date: | 2019-03-02 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | The mechanism of RNA duplex recognition and unwinding by DEAD-box helicase DDX3X.
Nat Commun, 10, 2019
|
|
6QDV
 
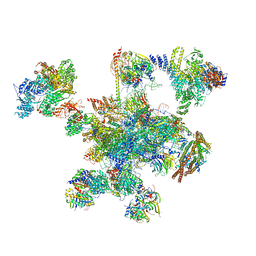 | | Human post-catalytic P complex spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Fica, S.M, Oubridge, C, Wilkinson, M.E, Newman, A.J, Nagai, K. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation.
Science, 363, 2019
|
|
6QIC
 
 | |
6QID
 
 | | Crystal structure of DEAH-box ATPase Prp43-S387A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Hamann, F, Ficner, R, Enders, M. | | Deposit date: | 2019-01-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis for RNA translocation by DEAH-box ATPases.
Nucleic Acids Res., 47, 2019
|
|
6QIE
 
 | | Crystal structure of DEAH-box ATPase Prp43-S387G | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Hamann, F, Ficner, R, Enders, M. | | Deposit date: | 2019-01-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for RNA translocation by DEAH-box ATPases.
Nucleic Acids Res., 47, 2019
|
|
6QV3
 
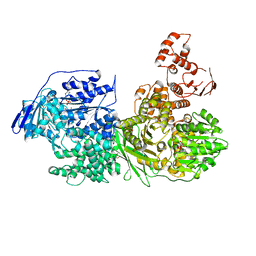 | | Crystal structure of the Ski2 RNA-helicase Brr2 from Chaetomium thermophilum bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Absmeier, E, Santos, K.F, Wahl, M.C. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Mechanism Underlying Inhibition of Intrinsic ATPase Activity in a Ski2-like RNA Helicase.
Structure, 28, 2020
|
|
6QV4
 
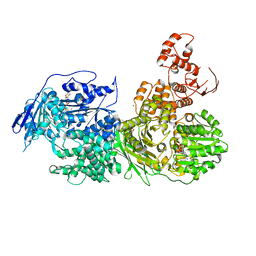 | | Crystal structure of the Ski2 RNA-helicase Brr2 from Chaetomium thermophilum bound to ATP-gamma-S | | Descriptor: | ACETATE ION, MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Absmeier, E, Santos, K.F, Wahl, M.C. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Mechanism Underlying Inhibition of Intrinsic ATPase Activity in a Ski2-like RNA Helicase.
Structure, 28, 2020
|
|
6QW6
 
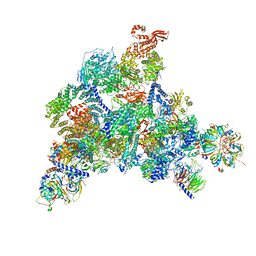 | | Structure of the human U5.U4/U6 tri-snRNP at 2.9A resolution. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Charenton, C, Wilkinson, M.E, Nagai, K. | | Deposit date: | 2019-03-05 | | Release date: | 2019-04-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Mechanism of 5' splice site transfer for human spliceosome activation.
Science, 364, 2019
|
|
6QWS
 
 | |
6QX9
 
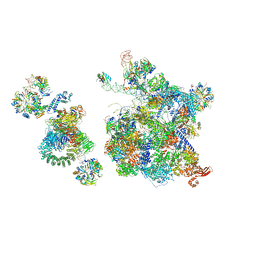 | | Structure of a human fully-assembled precatalytic spliceosome (pre-B complex). | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, AdML pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Charenton, C, Wilkinson, M.E, Nagai, K. | | Deposit date: | 2019-03-07 | | Release date: | 2019-04-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Mechanism of 5' splice site transfer for human spliceosome activation.
Science, 364, 2019
|
|
6RM8
 
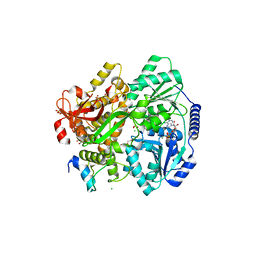 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2019-05-06 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RM9
 
 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2019-05-06 | | Release date: | 2020-02-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
