1A1O
 
 | | MHC CLASS I MOLECULE B*5301 COMPLEXED WITH PEPTIDE LS6 (KPIVQYDNF) FROM THE MALARIA PARASITE P. FALCIPARUM | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Harlos, K, Mcmichael, A.J, Stuart, D.I, Bell, J.I, Jones, E.Y. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53.
Immunity, 4, 1996
|
|
1A1P
 
 | | COMPSTATIN, NMR, 21 STRUCTURES | | Descriptor: | COMPSTATIN | | Authors: | Morikis, D, Assa-Munt, N, Sahu, A, Lambris, J.D. | | Deposit date: | 1997-12-12 | | Release date: | 1998-04-08 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Compstatin, a potent complement inhibitor.
Protein Sci., 7, 1998
|
|
1A1Q
 
 | | HEPATITIS C VIRUS NS3 PROTEINASE | | Descriptor: | NS3 PROTEINASE, ZINC ION | | Authors: | Love, R.A, Parge, H.E, Wickersham, J.A, Hostomsky, Z, Habuka, N, Moomaw, E.W, Adachi, T, Hostomska, Z. | | Deposit date: | 1997-12-12 | | Release date: | 1998-03-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site.
Cell(Cambridge,Mass.), 87, 1996
|
|
1A1R
 
 | | HCV NS3 PROTEASE DOMAIN:NS4A PEPTIDE COMPLEX | | Descriptor: | NS3 PROTEIN, NS4A PROTEIN, ZINC ION | | Authors: | Kim, J.L, Morgenstern, K.A, Lin, C, Fox, T, Dwyer, M.D, Landro, J.A, Chambers, S.P, Markland, W, Lepre, C.A, O'Malley, E.T, Harbeson, S.L, Rice, C.M, Murcko, M.A, Caron, P.R, Thomson, J.A. | | Deposit date: | 1997-12-15 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide.
Cell(Cambridge,Mass.), 87, 1996
|
|
1A1S
 
 | | ORNITHINE CARBAMOYLTRANSFERASE FROM PYROCOCCUS FURIOSUS | | Descriptor: | ORNITHINE CARBAMOYLTRANSFERASE | | Authors: | Villeret, V, Clantin, B, Tricot, C, Legrain, C, Roovers, M, Stalon, V, Glansdorff, N, Van Beeumen, J. | | Deposit date: | 1997-12-15 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of Pyrococcus furiosus ornithine carbamoyltransferase reveals a key role for oligomerization in enzyme stability at extremely high temperatures.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1A1T
 
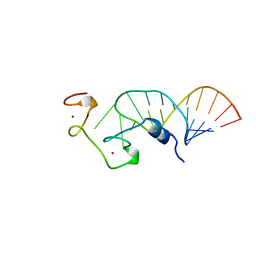 | | STRUCTURE OF THE HIV-1 NUCLEOCAPSID PROTEIN BOUND TO THE SL3 PSI-RNA RECOGNITION ELEMENT, NMR, 25 STRUCTURES | | Descriptor: | NUCLEOCAPSID PROTEIN, SL3 STEM-LOOP RNA, ZINC ION | | Authors: | De Guzman, R.N, Wu, Z.R, Stalling, C.C, Pappalardo, L, Borer, P.N, Summers, M.F. | | Deposit date: | 1997-12-15 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element.
Science, 279, 1998
|
|
1A1U
 
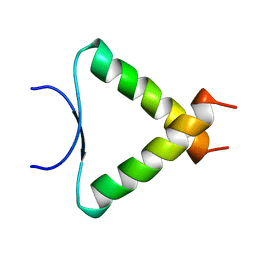 | | SOLUTION STRUCTURE DETERMINATION OF A P53 MUTANT DIMERIZATION DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | P53 | | Authors: | Mccoy, M.A, Stavridi, E.S, Waterman, J.L.F, Wieczorek, A, Opella, S.J, Halezonetis, T.D. | | Deposit date: | 1997-12-16 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Hydrophobic side-chain size is a determinant of the three-dimensional structure of the p53 oligomerization domain.
EMBO J., 16, 1997
|
|
1A1V
 
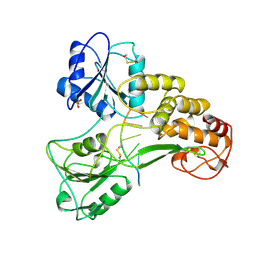 | | HEPATITIS C VIRUS NS3 HELICASE DOMAIN COMPLEXED WITH SINGLE STRANDED SDNA | | Descriptor: | DNA (5'-D(*UP*UP*UP*UP*UP*UP*UP*U)-3'), PROTEIN (NS3 PROTEIN), SULFATE ION | | Authors: | Kim, J.L, Morgenstern, K.A, Griffith, J.P, Dwyer, M.D, Thomson, J.A, Murcko, M.A, Lin, C, Caron, P.R. | | Deposit date: | 1997-12-17 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding.
Structure, 6, 1998
|
|
1A1W
 
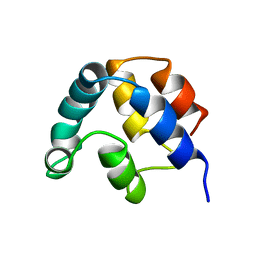 | | FADD DEATH EFFECTOR DOMAIN, F25Y MUTANT, NMR MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FADD PROTEIN | | Authors: | Eberstadt, M, Huang, B, Chen, Z, Meadows, R.P, Ng, C, Fesik, S.W. | | Deposit date: | 1997-12-18 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and mutagenesis of the FADD (Mort1) death-effector domain.
Nature, 392, 1998
|
|
1A1X
 
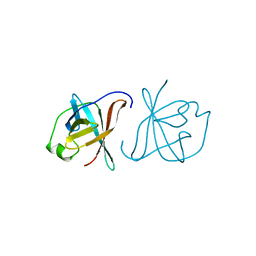 | | CRYSTAL STRUCTURE OF MTCP-1 INVOLVED IN T CELL MALIGNANCIES | | Descriptor: | HMTCP-1 | | Authors: | Fu, Z.Q, Dubois, G.C, Song, S.P, Kulikovskaya, I, Virgilio, L, Rothstein, J, Croce, C.M, Weber, I.T, Harrison, R.W. | | Deposit date: | 1997-12-18 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MTCP-1: implications for role of TCL-1 and MTCP-1 in T cell malignancies.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1A1Z
 
 | | FADD DEATH EFFECTOR DOMAIN, F25G MUTANT, NMR MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FADD PROTEIN | | Authors: | Eberstadt, M, Huang, B, Chen, Z, Meadows, R.P, Ng, C, Fesik, S.W. | | Deposit date: | 1997-12-18 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and mutagenesis of the FADD (Mort1) death-effector domain.
Nature, 392, 1998
|
|
1A21
 
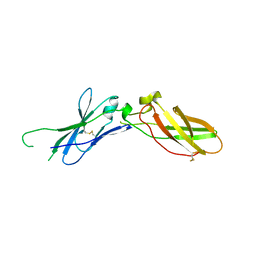 | | TISSUE FACTOR (TF) FROM RABBIT | | Descriptor: | TISSUE FACTOR | | Authors: | Muller, Y.A, De Vos, A.M. | | Deposit date: | 1998-01-14 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Hinge bending within the cytokine receptor superfamily revealed by the 2.4 A crystal structure of the extracellular domain of rabbit tissue factor.
Protein Sci., 7, 1998
|
|
1A22
 
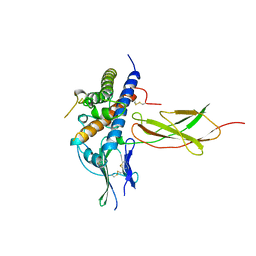 | | HUMAN GROWTH HORMONE BOUND TO SINGLE RECEPTOR | | Descriptor: | GROWTH HORMONE, GROWTH HORMONE RECEPTOR | | Authors: | De Vos, A.M, Ultsch, M. | | Deposit date: | 1998-01-15 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional analysis of the 1:1 growth hormone:receptor complex reveals the molecular basis for receptor affinity.
J.Mol.Biol., 277, 1998
|
|
1A23
 
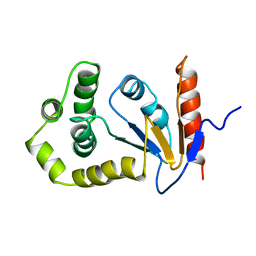 | | SOLUTION NMR STRUCTURE OF REDUCED DSBA FROM ESCHERICHIA COLI, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DSBA | | Authors: | Schirra, H.J, Renner, C, Czisch, M, Huber-Wunderlich, M, Holak, T.A, Glockshuber, R. | | Deposit date: | 1998-01-15 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of reduced DsbA from Escherichia coli in solution.
Biochemistry, 37, 1998
|
|
1A24
 
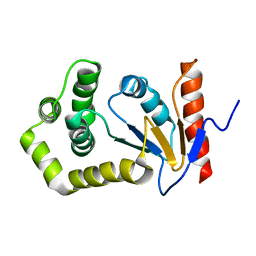 | | SOLUTION NMR STRUCTURE OF REDUCED DSBA FROM ESCHERICHIA COLI, FAMILY OF 20 STRUCTURES | | Descriptor: | DSBA | | Authors: | Schirra, H.J, Renner, C, Czisch, M, Huber-Wunderlich, M, Holak, T.A, Glockshuber, R. | | Deposit date: | 1998-01-15 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of reduced DsbA from Escherichia coli in solution.
Biochemistry, 37, 1998
|
|
1A25
 
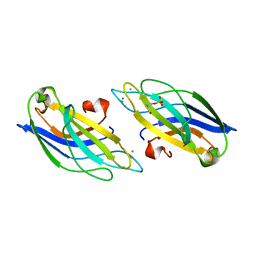 | | C2 DOMAIN FROM PROTEIN KINASE C (BETA) | | Descriptor: | CALCIUM ION, O-PHOSPHOETHANOLAMINE, PROTEIN KINASE C (BETA) | | Authors: | Sutton, R.B, Sprang, S.R. | | Deposit date: | 1998-01-16 | | Release date: | 1998-05-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the protein kinase Cbeta phospholipid-binding C2 domain complexed with Ca2+.
Structure, 6, 1998
|
|
1A26
 
 | |
1A27
 
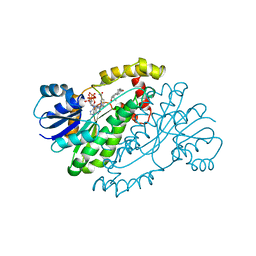 | | HUMAN 17-BETA-HYDROXYSTEROID-DEHYDROGENASE TYPE 1 C-TERMINAL DELETION MUTANT COMPLEXED WITH ESTRADIOL AND NADP+ | | Descriptor: | 17-BETA-HYDROXYSTEROID-DEHYDROGENASE, ESTRADIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mazza, C, Breton, R, Housset, D, Fontecilla-Camps, J.-C. | | Deposit date: | 1998-01-16 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human Type I 17Beta-Hydroxysteroid Dehydrogenase: Site Directed Mutagenesis and X-Ray Crystallography Structure-Function Analysis
Thesis, Universite Joseph Fourier, 1997
|
|
1A28
 
 | |
1A29
 
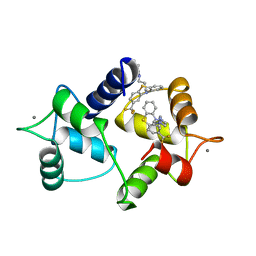 | | CALMODULIN COMPLEXED WITH TRIFLUOPERAZINE (1:2 COMPLEX) | | Descriptor: | 10-[3-(4-METHYL-PIPERAZIN-1-YL)-PROPYL]-2-TRIFLUOROMETHYL-10H-PHENOTHIAZINE, CALCIUM ION, CALMODULIN | | Authors: | Bocskei, Zs, Harmat, V, Vertessy, B.G, Ovadi, J, Naray-Szabo, G. | | Deposit date: | 1998-01-19 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Simultaneous binding of drugs with different chemical structures to Ca2+-calmodulin: crystallographic and spectroscopic studies.
Biochemistry, 37, 1998
|
|
1A2A
 
 | | AGKISTROTOXIN, A PHOSPHOLIPASE A2-TYPE PRESYNAPTIC NEUROTOXIN FROM AGKISTRODON HALYS PALLAS | | Descriptor: | CHLORIDE ION, PHOSPHOLIPASE A2 | | Authors: | Tang, L, Zhou, Y, Lin, Z. | | Deposit date: | 1997-12-25 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of agkistrodotoxin, a phospholipase A2-type presynaptic neurotoxin from agkistrodon halys pallas.
J.Mol.Biol., 282, 1998
|
|
1A2B
 
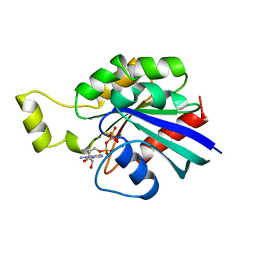 | | HUMAN RHOA COMPLEXED WITH GTP ANALOGUE | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, TRANSFORMING PROTEIN RHOA | | Authors: | Ihara, K, Muraguchi, S, Kato, M, Shimizu, T, Shirakawa, M, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 1997-12-26 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue.
J.Biol.Chem., 273, 1998
|
|
1A2C
 
 | | Structure of thrombin inhibited by AERUGINOSIN298-A from a BLUE-GREEN ALGA | | Descriptor: | Aeruginosin 298-A, Hirudin variant-2, SODIUM ION, ... | | Authors: | Rios-Steiner, J.L, Murakami, M, Tulinsky, A. | | Deposit date: | 1997-12-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Thrombin Inhibited by Aeruginosin 298-A from a Blue-Green Alga
J.Am.Chem.Soc., 120, 1998
|
|
1A2D
 
 | | PYRIDOXAMINE MODIFIED MURINE ADIPOCYTE LIPID BINDING PROTEIN | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN, CHLORIDE ION | | Authors: | Ory, J, Mazhary, A, Kuang, H, Davies, R, Distefano, M, Banaszak, L. | | Deposit date: | 1997-12-29 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of two synthetic catalysts based on adipocyte lipid-binding protein.
Protein Eng., 11, 1998
|
|
1A2E
 
 | |
