5NEV
 
 | | CDK2/Cyclin A in complex with compound 73 | | Descriptor: | 4-[[6-(3-phenylphenyl)-7~{H}-purin-2-yl]amino]benzenesulfonamide, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E.M, Newell, D.R, Turner, D, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Cano, G. | | Deposit date: | 2017-03-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
5N01
 
 | | Crystal structure of the decarboxylase AibA/AibB C56N variant | | Descriptor: | ACETATE ION, GLYCEROL, Glutaconate CoA-transferase family, ... | | Authors: | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6S32
 
 | | Crystal structure of ene-reductase CtOYE from Chroococcidiopsis thermalis. | | Descriptor: | ACETATE ION, BENZAMIDINE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Robescu, M.R, Niero, M, Hall, M, Bergantino, E, Cendron, L. | | Deposit date: | 2019-06-24 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Two new ene-reductases from photosynthetic extremophiles enlarge the panel of old yellow enzymes: CtOYE and GsOYE.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
6RX1
 
 | | Crystal structure of human syncytin 1 in post-fusion conformation | | Descriptor: | CHLORIDE ION, GLYCEROL, Syncytin-1 | | Authors: | Ruigrok, K, Backovic, M, Vaney, M.C, Rey, F.A. | | Deposit date: | 2019-06-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structures of the Post-fusion 6-Helix Bundle of the Human Syncytins and their Functional Implications.
J.Mol.Biol., 431, 2019
|
|
5N1Y
 
 | | HLA-A02 carrying MVWGPDPLYV | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2017-02-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J. Clin. Invest., 126, 2016
|
|
6S73
 
 | | Crystal structure of Nek7 SRS mutant bound to compound 51 | | Descriptor: | 3-[[6-(cyclohexylmethoxy)-7~{H}-purin-2-yl]amino]-~{N}-[3-(dimethylamino)propyl]benzenesulfonamide, Serine/threonine-protein kinase Nek7 | | Authors: | Nasir, N, Byrne, M.J, Bhatia, C, Bayliss, R. | | Deposit date: | 2019-07-04 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nek7 conformational flexibility and inhibitor binding probed through protein engineering of the R-spine.
Biochem.J., 477, 2020
|
|
5N2L
 
 | | Crystal structure of the second bromodomain of human BRD2 in complex with a tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, SULFATE ION, ... | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Wiggers, H.J, Picaud, S, Fedorov, O, Krojer, T, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the second bromodomain of human BRD2 in complex with a tetrahydroquinoline analogue
To Be Published
|
|
6RX2
 
 | | Fragment AZ-005 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 7-[[(2~{S})-1-azanylpropan-2-yl]amino]-1-benzothiophene-2-carboximidamide, CALCIUM ION, ... | | Authors: | Leysen, S, Guillory, X, Wolter, M, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-06-07 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
5NNH
 
 | | KSHV uracil-DNA glycosylase, apo form | | Descriptor: | SULFATE ION, Uracil-DNA glycosylase | | Authors: | Earl, C, Bagneris, C, Cole, A.R, Barrett, T, Savva, R. | | Deposit date: | 2017-04-09 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Nucleic Acids Res., 46, 2018
|
|
6S77
 
 | | Crystal structure of CARM1 N265Y mutant in complex with inhibitor AA183 | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-(pyridin-2-ylamino)propyl]amino]-2-azanyl-butanoic acid, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
6S7Y
 
 | |
6S82
 
 | | Structure Of D80A-Fructofuranosidase From Xanthophyllomyces Dendrorhous Complexed With Tris-buffer molecule And hydroquinone | | Descriptor: | 1,2-ETHANEDIOL, 1,4-benzoquinone, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2019-07-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Deciphering the molecular specificity of phenolic compounds as inhibitors or glycosyl acceptors of beta-fructofuranosidase from Xanthophyllomyces dendrorhous.
Sci Rep, 9, 2019
|
|
5N38
 
 | | S65DParkin and pUB complex | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6RFC
 
 | | Crystal structure of the potassium-pumping G263F mutant of the light-driven sodium pump KR2 in the monomeric form, pH 4.3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, GLYCEROL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RGF
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with an inhibitor | | Descriptor: | CITRIC ACID, NAD kinase 1, [(2~{R},3~{R},4~{R},5~{R})-2-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-methyl-amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-5-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl] dihydrogen phosphate | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with an inhibitor
To Be Published
|
|
6RX7
 
 | |
5NPU
 
 | | Inferred ancestral pyruvate decarboxylase | | Descriptor: | ANC27, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Buddrus, L, Crennell, S.J, Leak, D.J, Danson, M.J, Andrews, E.S.V, Arcus, V.L. | | Deposit date: | 2017-04-19 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of an inferred ancestral bacterial pyruvate decarboxylase.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6RXB
 
 | | Crystal structure of TetR-Q116A from Acinetobacter baumannii AYE in complex with minocycline | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Tam, H.K, Himpich, S, Pos, K.M. | | Deposit date: | 2019-06-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Binding of Tetracyclines to Acinetobacter baumannii TetR Involves Two Arginines as Specificity Determinants
Front Microbiol, 2021
|
|
6RHC
 
 | | Fragment AZ-003 binding at the TAZpS89/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-azanyl-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
5N9K
 
 | | Crystal structure of human Protein kinase CK2 catalytic subunit in complex with the ATP-competitive, tight-binding dibenzofuran inhibitor TF107 (5) | | Descriptor: | 1,3-bis(chloranyl)-6-[(~{E})-(4-methoxyphenyl)iminomethyl]dibenzofuran-2,7-diol, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Schnitzler, A, Gratz, A, Bollacke, A, Weyrich, M, Kucklaender, U, Wuensch, B, Goetz, C, Niefind, K, Jose, J. | | Deposit date: | 2017-02-25 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.643 Å) | | Cite: | A pi-Halogen Bond of Dibenzofuranones with the Gatekeeper Phe113 in Human Protein Kinase CK2 Leads to Potent Tight Binding Inhibitors.
Pharmaceuticals, 11, 2018
|
|
6S1R
 
 | |
5N9N
 
 | | Crystal structure of human Protein kinase CK2 catalytic subunit in complex with the ATP-competitive, tight-binding dibenzofuran inhibitor TF85 (4a) | | Descriptor: | (4~{Z})-7,9-bis(chloranyl)-4-[[(4-methoxyphenyl)amino]methylidene]-8-oxidanyl-1,2-dihydrodibenzofuran-3-one, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Schnitzler, A, Gratz, A, Bollacke, A, Weyrich, M, Kucklaender, U, Wuensch, B, Goetz, C, Niefind, K, Jose, J. | | Deposit date: | 2017-02-25 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | A pi-Halogen Bond of Dibenzofuranones with the Gatekeeper Phe113 in Human Protein Kinase CK2 Leads to Potent Tight Binding Inhibitors.
Pharmaceuticals, 11, 2018
|
|
6S1X
 
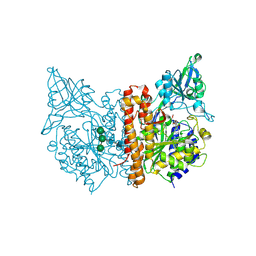 | |
5NAD
 
 | | TTK kinase domain in complex with BAY 1217389 | | Descriptor: | BAY 1217389, Dual specificity protein kinase TTK | | Authors: | Uitdehaag, J.C.M, Willemsen-Seegers, N, Sterrenburg, J.G, de Man, J, Buijsman, R.C, Zaman, G.J.R. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Target Residence Time-Guided Optimization on TTK Kinase Results in Inhibitors with Potent Anti-Proliferative Activity.
J. Mol. Biol., 429, 2017
|
|
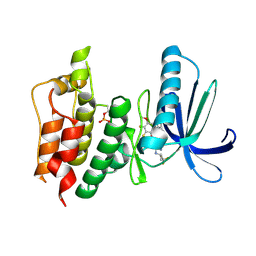
6RKI
 
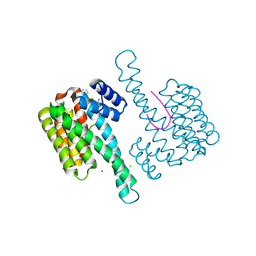 | | Fragment AZ-023 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-[(3-aminophenyl)amino]-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
