6AH0
 
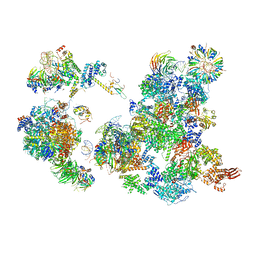 | | The Cryo-EM Structure of the Precusor of Human Pre-catalytic Spliceosome (pre-B complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | Deposit date: | 2018-08-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
4QSP
 
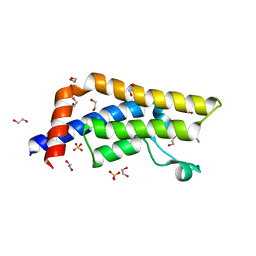 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with acetyl-lysine | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, N(6)-ACETYLLYSINE, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
4QSV
 
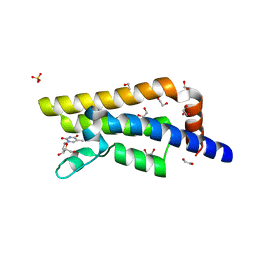 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with thymidine | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, SULFATE ION, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
4QTU
 
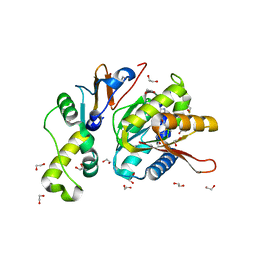 | | Structure of S. cerevisiae Bud23-Trm112 complex involved in formation of m7G1575 on 18S rRNA (SAM bound form) | | Descriptor: | 1,2-ETHANEDIOL, Multifunctional methyltransferase subunit TRM112, Putative methyltransferase BUD23, ... | | Authors: | Letoquart, J, Huvelle, E, Wacheul, L, Bourgeois, G, Zorbas, C, Graille, M, Heurgue-Hamard, V, Lafontaine, D.L.J. | | Deposit date: | 2014-07-09 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.124 Å) | | Cite: | Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QSQ
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) with bound DMSO | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
4QSW
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with 5-methyl uridine | | Descriptor: | 1,2-ETHANEDIOL, 5-methyluridine, ATPase family AAA domain-containing protein 2, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
1OSV
 
 | | STRUCTURAL BASIS FOR BILE ACID BINDING AND ACTIVATION OF THE NUCLEAR RECEPTOR FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile acid receptor, Nuclear receptor coactivator 2 | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
1OVE
 
 | | The structure of p38 alpha in complex with a dihydroquinolinone | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-5-(2,4-DIFLUOROPHENYL)-7-PIPERIDIN-4-YL-3,4-DIHYDROQUINOLIN-2(1H)-ONE, GLYCEROL, Mitogen-activated protein kinase 14 | | Authors: | Fitzgerald, C.E, Patel, S.B, Becker, J.W, Cameron, P.M, Zaller, D, Pikounis, V.B, O'Keefe, S.J, Scapin, G. | | Deposit date: | 2003-03-26 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for p38alpha MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity
Nat.Struct.Biol., 10, 2003
|
|
2LCW
 
 | |
1OUK
 
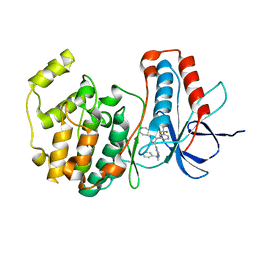 | | The structure of p38 alpha in complex with a pyridinylimidazole inhibitor | | Descriptor: | 4-[5-[2-(1-PHENYL-ETHYLAMINO)-PYRIMIDIN-4-YL]-1-METHYL-4-(3-TRIFLUOROMETHYLPHENYL)-1H-IMIDAZOL-2-YL]-PIPERIDINE, Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Fitzgerald, C.E, Patel, S.B, Becker, J.W, Cameron, P.M, Zaller, D, Pikounis, V.B, O'Keefe, S.J, Scapin, G. | | Deposit date: | 2003-03-24 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for p38alpha MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity
Nat.Struct.Biol., 10, 2003
|
|
1OUY
 
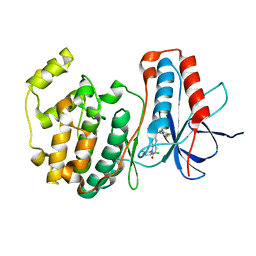 | | The structure of p38 alpha in complex with a dihydropyrido-pyrimidine inhibitor | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-6-[(2,4-DIFLUOROPHENYL)SULFANYL]-7-(1,2,3,6-TETRAHYDRO-4-PYRIDINYL)-3,4-DIHYDROPYRIDO[3,2-D]PYRIMIDIN-2(1H)-ONE, Mitogen-activated protein kinase 14 | | Authors: | Fitzgerald, C.E, Patel, S.B, Becker, J.W, Cameron, P.M, Zaller, D, Pikounis, V.B, O'Keefe, S.J, Scapin, G. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for p38alpha MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity
Nat.Struct.Biol., 10, 2003
|
|
4R70
 
 | |
4ITE
 
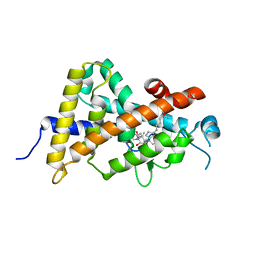 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 1alpha,25-Dihydroxy-2alpha-[2-(2H-tetrazol-2-yl)ethyl]vitamin D3 | | Descriptor: | (1R,2S,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-7a-methyl-1-[(2R)-6-methyl-6-oxidanyl-heptan-2-yl]-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-2-[2-(1,2,3,4-tetrazol-2-yl)ethyl]cyclohexane-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2013-01-18 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Synthesis of 2 alpha-heteroarylalkyl active vitamin d3 with therapeutic effect on enhancing bone mineral density in vivo
ACS MED.CHEM.LETT., 4, 2013
|
|
3S3I
 
 | | p38 kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | 3-(3-tert-butyl[1,2,4]triazolo[4,3-a]pyridin-7-yl)-N-cyclopropyl-4-methylbenzamide, Mitogen-activated protein kinase 14 | | Authors: | Segarra, V, Aiguade, J, Roca, R, Fisher, M, Lamers, M. | | Deposit date: | 2011-05-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel triazolopyridylbenzamides as potent and selective p38 alpha inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4RAG
 
 | | CRYSTAL STRUCTURE of PPC2A-D38K | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 1A | | Authors: | Pan, C, Tang, J.Y, Xu, Y.F, Xiao, P, Liu, H.D, Wang, H.A, Wang, W.B, Meng, F.G, Yu, X, Sun, J.P. | | Deposit date: | 2014-09-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The catalytic role of the M2 metal ion in PP2C alpha
Sci Rep, 5, 2015
|
|
8AO8
 
 | | Specific covalent inhibitor(9) of ERK2 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2~{R})-2-(3-methylimidazol-4-yl)piperidin-1-yl]propan-1-one, Mitogen-activated protein kinase 1, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
8AOB
 
 | | Specific covalent inhibitor(12) of ERK2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Mitogen-activated protein kinase 1, SULFATE ION, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
8AO6
 
 | | electrophilic inhibitor (7) of ERK2 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Mitogen-activated protein kinase 1, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
8AOF
 
 | | Specific covalent inhibitor(16) of ERK2 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 1, SULFATE ION, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.615 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
8AO5
 
 | | Specific covalent inhibitor (6) of ERK2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Mitogen-activated protein kinase 1, SULFATE ION, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.595 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
8AO4
 
 | | Specific covalent inhibitor (5) of ERK2 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Mitogen-activated protein kinase 1, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.825 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
4R0E
 
 | |
8AOJ
 
 | | Specific covalent inhibitor of ERK2 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2~{S})-2-(5-methyl-3-pyridin-4-yl-1~{H}-pyrazol-4-yl)pyrrolidin-1-yl]propan-1-one, DIMETHYL SULFOXIDE, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
8AO3
 
 | | Specific covalent inhibitor of ERK2 | | Descriptor: | 2-chloranyl-~{N}-(1~{H}-indazol-5-ylmethyl)ethanamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
8AO9
 
 | | Specific covalent inhibitor(10) of ERK2 | | Descriptor: | 1,2-ETHANEDIOL, 1-(6,7-dihydro-4~{H}-[1,3]thiazolo[5,4-c]pyridin-5-yl)propan-1-one, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.624 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
