7XJ6
 
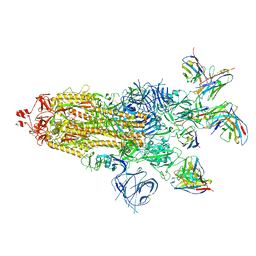 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 1 conformation
To Be Published
|
|
5UZE
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P182 | | Descriptor: | GLYCEROL, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-26 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P182
To Be Published
|
|
4BGQ
 
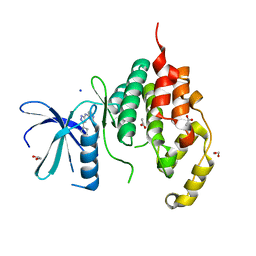 | | Crystal structure of the human CDKL5 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CYCLIN-DEPENDENT KINASE-LIKE 5, ... | | Authors: | Canning, P, Krojer, T, Goubin, S, Mahajan, P, Vollmar, M, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-03-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CDKL Family Kinases Have Evolved Distinct Structural Features and Ciliary Function.
Cell Rep, 22, 2018
|
|
3S4C
 
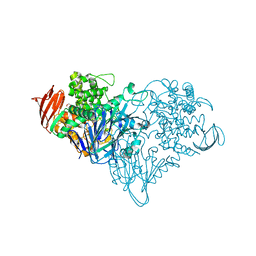 | | Lactose phosphorylase in complex with sulfate | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Lactose Phosphorylase, SULFATE ION | | Authors: | Van Hoorebeke, A, Stout, J, Soetaert, W, Van Beeumen, J, Desmet, T, Savvides, S. | | Deposit date: | 2011-05-19 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cellobiose phosphorylase: reconstructing the structural itinerary along the catalytic pathway
To be Published
|
|
4HVD
 
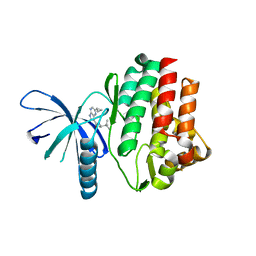 | | JAK3 kinase domain in complex with 2-Cyclopropyl-5H-pyrrolo[2,3-b]pyrazine-7-carboxylic acid ((S)-1,2,2-trimethyl-propyl)-amide | | Descriptor: | 1-phenylurea, 2-cyclopropyl-N-[(2S)-3,3-dimethylbutan-2-yl]-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Kuglstatter, A, Shao, A. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 3-Amido Pyrrolopyrazine JAK Kinase Inhibitors: Development of a JAK3 vs JAK1 Selective Inhibitor and Evaluation in Cellular and in Vivo Models.
J.Med.Chem., 56, 2013
|
|
2XUC
 
 | | Natural product-guided discovery of a fungal chitinase inhibitor | | Descriptor: | 1-methyl-3-(N-methylcarbamimidoyl)urea, CHITINASE, CHLORIDE ION, ... | | Authors: | Rush, C.L, Schuttelkopf, A.W, Hurtado-Guerrero, R, Blair, D.E, Ibrahim, A.F.M, Desvergnes, S, Eggleston, I.M, van Aalten, D.M.F. | | Deposit date: | 2010-10-18 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Natural Product-Guided Discovery of a Fungal Chitinase Inhibitor.
Chem.Biol., 17, 2010
|
|
4B5U
 
 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with pyruvate and succinic semialdehyde | | Descriptor: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, 4-oxobutanoic acid, COBALT (II) ION, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.913 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
3E73
 
 | | Crystal Structure of Human LanCL1 complexed with GSH | | Descriptor: | GLUTATHIONE, LanC-like protein 1, ZINC ION | | Authors: | Zhang, W, Zhu, G, Li, X, Rao, Z, Zhang, C. | | Deposit date: | 2008-08-17 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione
Genes Dev., 23, 2009
|
|
4B5T
 
 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with ketobutyrate | | Descriptor: | 2-KETOBUTYRIC ACID, 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, COBALT (II) ION, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
5ZCM
 
 | |
4B8Y
 
 | | Ferrichrome-bound FhuD2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Malito, E, Bottomley, M.J, Spraggon, G. | | Deposit date: | 2012-08-31 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of the Staphylococcus Aureus Virulence Factor and Vaccine Candidate Fhud2.
Biochem.J., 449, 2013
|
|
4I9T
 
 | | Structure of the H258Y mutant of the phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, SULFATE ION, ... | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4QJW
 
 | | Crystal structure of catalytic domain of human carbonic anhydrase isozyme XII with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-(benzylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-06-05 | | Release date: | 2015-04-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Functionalization of Fluorinated Benzenesulfonamides and Their Inhibitory Properties toward Carbonic Anhydrases
Chemmedchem, 10, 2015
|
|
3E6U
 
 | | Crystal structure of Human LanCL1 | | Descriptor: | LanC-like protein 1, ZINC ION | | Authors: | Zhang, W, Zhu, G, Li, X, Rao, Z, Zhang, C. | | Deposit date: | 2008-08-16 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione
Genes Dev., 23, 2009
|
|
3GVD
 
 | | Crystal Structure of Serine Acetyltransferase CysE from Yersinia pestis | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ACETIC ACID, CYSTEINE, ... | | Authors: | Kim, Y, Zhou, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-30 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Serine Acetyltransferase CysE from Yersinia pestis
To be Published
|
|
6ECH
 
 | | Pyruvate Kinase Isoform L-type with phosphorylated Ser12 (pS12) in complex with FBP | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Padyana, A, Tong, S. | | Deposit date: | 2018-08-07 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Distinct Hepatic PKA and CDK Signaling Pathways Control Activity-Independent Pyruvate Kinase Phosphorylation and Hepatic Glucose Production.
Cell Rep, 29, 2019
|
|
5LA4
 
 | | Crystal structure of apo human proheparanase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heparanase, ... | | Authors: | Wu, L, Jin, Y, Davies, G.J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
7K6Z
 
 | | Carbonic Anhydrase IX mimic complexed with 4-(2-(3-(4-fluorophenyl)ureido)ethylsulfonamido)benzenesulfonamide | | Descriptor: | 4-{[(2-{[(4-fluorophenyl)carbamoyl]amino}ethyl)sulfonyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Andring, J.T, Singh, S, McKenna, R. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | Handling drug-target selectivity: A study on ureido containing Carbonic Anhydrase inhibitors.
Eur.J.Med.Chem., 212, 2021
|
|
1FCA
 
 | |
3NJU
 
 | | Crystal structure of the complex of group I phospholipase A2 with 4-Methoxy-benzoicacid at 1.4A resolution | | Descriptor: | 4-METHOXYBENZOIC ACID, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Kaushik, S, Prem Kumar, R, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the complex of group I phospholipase A2 with 4-Methoxy-benzoicacid at 1.4A resolution
To be Published
|
|
1ZDX
 
 | | Solution Structure of the type 1 pilus assembly platform FimD(25-125) | | Descriptor: | Outer membrane usher protein fimD | | Authors: | Nishiyama, M, Horst, R, Herrmann, T, Vetsch, M, Bettendorff, P, Ignatov, O, Grutter, M, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
4QML
 
 | | MST3 in complex with AMP-PNP | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Olesen, S.H, Watts, C, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Discovery of Diverse Small-Molecule Inhibitors of Mammalian Sterile20-like Kinase 3 (MST3).
Chemmedchem, 11, 2016
|
|
3HCA
 
 | | Crystal Structure of E185Q hPNMT in Complex With Octopamine and AdoHcy | | Descriptor: | 1,2-ETHANEDIOL, 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, Phenylethanolamine N-methyltransferase, ... | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
Biochem.J., 422, 2009
|
|
3K14
 
 | | Co-crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with FOL fragment 535, ethyl 3-methyl-5,6-dihydroimidazo[2,1-b][1,3]thiazole-2-carboxylate | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
5VAU
 
 | |
