4U32
 
 | | Human mesotrypsin complexed with HAI-2 Kunitz domain 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Kunitz-type protease inhibitor 2, ... | | Authors: | Wang, R, Soares, A.S, Radisky, E.S. | | Deposit date: | 2014-07-18 | | Release date: | 2014-10-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Sequence and Conformational Specificity in Substrate Recognition: SEVERAL HUMAN KUNITZ PROTEASE INHIBITOR DOMAINS ARE SPECIFIC SUBSTRATES OF MESOTRYPSIN.
J.Biol.Chem., 289, 2014
|
|
1U2N
 
 | | Structure CBP TAZ1 Domain | | Descriptor: | CREB binding protein, ZINC ION | | Authors: | De Guzman, R.N, Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-07-19 | | Release date: | 2005-04-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding
Biochemistry, 44, 2005
|
|
8AK4
 
 | | Structure of the C-terminally truncated NAD+-dependent DNA ligase from the poly-extremophile Deinococcus radiodurans | | Descriptor: | DNA ligase, MANGANESE (II) ION, ZINC ION | | Authors: | Fernandes, A, Williamson, A.K, Matias, P.M, Moe, E. | | Deposit date: | 2022-07-29 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structure/function studies of the NAD + -dependent DNA ligase from the poly-extremophile Deinococcus radiodurans reveal importance of the BRCT domain for DNA binding.
Extremophiles, 27, 2023
|
|
8AW3
 
 | | Cryo-EM structure of the Tb ADAT2/3 deaminase in complex with tRNA | | Descriptor: | Deaminase, putative, RNA (75-MER), ... | | Authors: | Dolce, L.G, Tengo, L, Weis, F, Kowalinski, E. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for sequence-independent substrate selection by eukaryotic wobble base tRNA deaminase ADAT2/3.
Nat Commun, 13, 2022
|
|
4V8M
 
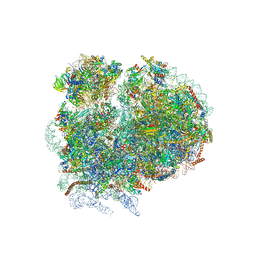 | | High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome | | Descriptor: | 18S RRNA OF THE SMALL RIBOSOMAL SUBUNIT, 40S RIBOSOMAL PROTEIN S10, PUTATIVE, ... | | Authors: | Hashem, Y, des Georges, A, Fu, J, Buss, S.N, Jossinet, F, Jobe, A, Zhang, Q, Liao, H.Y, Grassucci, R.A, Bajaj, C, Westhof, E, Madison-Antenucci, S, Frank, J. | | Deposit date: | 2012-12-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.57 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure of the Trypanosoma Brucei Ribosome.
Nature, 494, 2013
|
|
5QHA
 
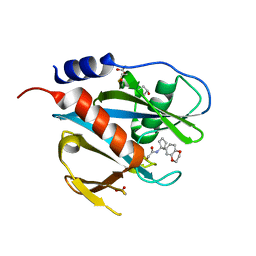 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with PCM-0102951 | | Descriptor: | ACETATE ION, N-[1-(2,3-dihydro-1,4-benzodioxin-6-yl)cyclopentyl]acetamide, Peroxisomal coenzyme A diphosphatase NUDT7 | | Authors: | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2018-09-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
9NJO
 
 | |
9NI0
 
 | |
9NAA
 
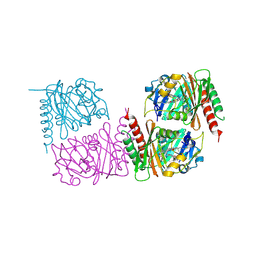 | | Unusual structure of a bacteriophytochrome fragment derived from full length SaBphP2 | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, histidine kinase | | Authors: | Schmidt, M, Prabin, K. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of myxobacterial phytochrome revealed by cryo-EM using the Spotiton technique and with x-ray crystallography.
Struct Dyn., 12, 2025
|
|
6KZA
 
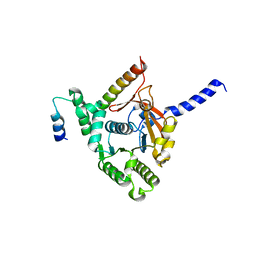 | | Crystal structure of the complex of the interaction domains of E. coli DnaB helicase and DnaC helicase loader | | Descriptor: | DNA replication protein DnaC, Replicative DNA helicase | | Authors: | Nagata, K, Okada, A, Ohtsuka, J, Ohkuri, T, Akama, Y, Sakiyama, Y, Miyazaki, E, Horita, S, Katayama, T, Ueda, T, Tanokura, M. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the complex of the interaction domains of Escherichia coli DnaB helicase and DnaC helicase loader: structural basis implying a distortion-accumulation mechanism for the DnaB ring opening caused by DnaC binding.
J.Biochem., 167, 2020
|
|
6LA8
 
 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 | | Descriptor: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Lee, P.L, Sharma, D, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
4UMC
 
 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase - the importance of accommodating the active site water | | Descriptor: | L-PHOSPHOLACTATE, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
6TCX
 
 | | Papain bound to a natural cysteine protease inhibitor from Streptomyces mobaraensis | | Descriptor: | (2~{R})-2-[[(1~{S})-1-[(6~{S})-2-azanyl-1,4,5,6-tetrahydropyrimidin-6-yl]-2-[[(2~{S})-3-methyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanyl-3-phenyl-propan-2-yl]amino]butan-2-yl]amino]-2-oxidanylidene-ethyl]carbamoylamino]-3-(4-hydroxyphenyl)propanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Papain | | Authors: | Kraemer, A, Juettner, N.E, Fuchsbauer, H.-L, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-06 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Decoding the Papain Inhibitor from Streptomyces mobaraensis as Being Hydroxylated Chymostatin Derivatives: Purification, Structure Analysis, and Putative Biosynthetic Pathway.
J.Nat.Prod., 83, 2020
|
|
9B4E
 
 | | Structure of wild type human PSS1 | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CALCIUM ION, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2024-03-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular insights into human phosphatidylserine synthase 1 reveal its inhibition promotes LDL uptake.
Cell, 187, 2024
|
|
9B4G
 
 | | Structure of inhibitor-bound human PSS1 | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, (7P)-7-[(4S)-4-{4-[3,5-bis(trifluoromethyl)phenoxy]phenyl}-5-(2,2-difluoropropyl)-6-oxo-1,4,5,6-tetrahydropyrrolo[3,4-c]pyrazol-3-yl]-1,3-benzoxazol-2(3H)-one, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2024-03-20 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Molecular insights into human phosphatidylserine synthase 1 reveal its inhibition promotes LDL uptake.
Cell, 187, 2024
|
|
5QCS
 
 | | Crystal structure of BACE complex with BMC024 | | Descriptor: | (2R,4S)-N-BUTYL-4-HYDROXY-2-METHYL- 4-((E)-(4AS,12R,15S,17AS)-15-METHYL -14,17-DIOXO-2,3,4,4A,6,9,11,12,13, 14,15,16,17,17A-TETRADECAHYDRO-1H-5 ,10-DITHIA-1,13,16-TRIAZA-BENZOCYCL OPENTADECEN-12-YL)-BUTYRAMIDE, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
4UMB
 
 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase - the importance of accommodating the active site water | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
6LA9
 
 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 (high cryoprotectant) | | Descriptor: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
4V14
 
 | |
8AVV
 
 | | Cryo-EM structure of DrBphP photosensory module in Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome,Response regulator | | Authors: | Wahlgren, W.Y, Takala, H, Westenhoff, S. | | Deposit date: | 2022-08-27 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of signal transduction in a phytochrome histidine kinase.
Nat Commun, 13, 2022
|
|
8AVW
 
 | | Cryo-EM structure of DrBphP in Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome,Response regulator | | Authors: | Wahlgren, W.Y, Takala, H, Westenhoff, S. | | Deposit date: | 2022-08-27 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural mechanism of signal transduction in a phytochrome histidine kinase.
Nat Commun, 13, 2022
|
|
5QD4
 
 | | Crystal structure of BACE complex with BMC023 | | Descriptor: | Beta-secretase 1, {(E)-(3R,6S,9R)-3-[(1S,3R)-3-((S)-1 -BUTYLCARBAMOYL-2-METHYL-PROPYLCARB AMOYL)-1-HYDROXY-BUTYL]-6-METHYL-5, 8-DIOXO-1,11-DITHIA-4,7-DIAZA-CYCLO PENTADEC-13-EN-9-YL}-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
9UHT
 
 | | SARS-CoV-2 E-RTC in complex with RNA-nsp9 and GMPPNP | | Descriptor: | Helicase nsp13, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Huang, Y.C, Liu, Y.X, Lou, Z.Y, Rao, Z.H, Yan, L.M. | | Deposit date: | 2025-04-14 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Response to Matter Arising: Substrate selection and transition state support the mechanism for GTP-mediate RNA capping catalyzed by SARS-CoV-2 NiRAN
To Be Published
|
|
7FIO
 
 | | LecA from Pseudomonas aeruginosa in complex with a synthetic monovalent galactosidic inhibitor | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-[2-[4-[(2S)-3-(2-hydroxyethylamino)-3-oxidanylidene-2-(2-phenoxyethanoylamino)propyl]-1,2,3-triazol-1-yl]ethyl]-4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-benzamide, ... | | Authors: | Kuhaudomlarp, S, Siebs, E, da Silva Figueiredo Celestino Gomes, P, Fortin, C, Rognan, D, Rademacher, C, Imberty, A, Titz, A. | | Deposit date: | 2021-07-31 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Targeting the Central Pocket of the Pseudomonas aeruginosa Lectin LecA.
Chembiochem, 23, 2022
|
|
9UDD
 
 | | Crystal structure of MonCI in complex with monoepoxidized farnesyl acetate | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Xiao, H.L, Guan, Y.Z, Chen, X. | | Deposit date: | 2025-04-06 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic and molecular insights into iterative triepoxidation catalyzed by monooxygenase MonCI using a natural substrate analog.
Int.J.Biol.Macromol., 311, 2025
|
|
