4NLB
 
 | |
5D9V
 
 | |
6GWS
 
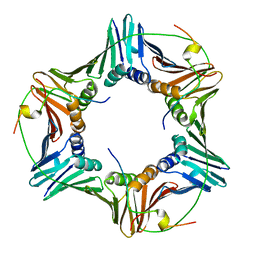 | | Crystal structure of human PCNA in complex with three p15 peptides | | Descriptor: | PCNA-associated factor, Proliferating cell nuclear antigen | | Authors: | De March, M, Merino, N, Gonzalez-Magana, A, Romano-Moreno, M, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2018-06-25 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | p15PAF binding to PCNA modulates the DNA sliding surface.
Nucleic Acids Res., 46, 2018
|
|
6C05
 
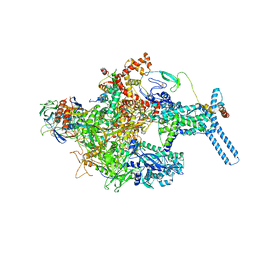 | | Mycobacterium tuberculosis RNAP Holo/RbpA in relaxed state | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J, Lilic, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-03-28 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (5.15 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
4NNJ
 
 | |
1HO5
 
 | |
6GHP
 
 | | 14-3-3sigma in complex with a TASK3 peptide stabilized by semi-synthetic natural product FC-NAc | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Potassium channel subfamily K member 9, ... | | Authors: | Andrei, S.A, de Vink, P.J, Brunsveld, L, Ottmann, C, Higuchi, Y. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-01 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rationally Designed Semisynthetic Natural Product Analogues for Stabilization of 14-3-3 Protein-Protein Interactions.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
1HQF
 
 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH N-HYDROXY-L-ARGININE | | Descriptor: | ARGINASE 1, MANGANESE (II) ION, N-OMEGA-HYDROXY-L-ARGININE | | Authors: | Cox, J.D, Cama, E, Colleluori, D.M, Ash, D.E, Christianson, D.W. | | Deposit date: | 2000-12-16 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic and metabolic inferences from the binding of substrate analogues and products to arginase.
Biochemistry, 40, 2001
|
|
1EUP
 
 | |
1E9V
 
 | | XENON BOUND IN HYDROPHOBIC CHANNEL OF HYBRID CLUSTER PROTEIN FROM DESULFOVIBRIO VULGARIS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Cooper, S.J, Bailey, S, Rizkallah, P.J, Lindley, P.F. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-25 | | Last modified: | 2020-12-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Ferricyanide Soaked Hybrid Cluster Protein at 1.2A and Xenon Mapping of the Hydrophobic Cavity at 1.8A
To be Published
|
|
1HJS
 
 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-1,4-GALACTANASE, ... | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
6H2T
 
 | | GlnH bound to Glu, Mycobacterium tuberculosis | | Descriptor: | GLUTAMIC ACID, Probable glutamine-binding lipoprotein GlnH (GLNBP) | | Authors: | O'Hare, H.M, Wallis, R, Lin, L.Y, Newland-Smith, Z. | | Deposit date: | 2018-07-16 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | An Aspartate-Specific Solute-Binding Protein Regulates Protein Kinase G Activity To Control Glutamate Metabolism in Mycobacteria.
MBio, 9, 2018
|
|
5D9B
 
 | | Luciferin-regenerating enzyme solved by SIRAS using XFEL (refined against native data) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
5CR1
 
 | | Crystal structure of TTR/resveratrol/T4 complex | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, RESVERATROL, Transthyretin | | Authors: | Zanotti, G, Florio, P, Folli, C, Cianci, M, Del Rio, D, Berni, R. | | Deposit date: | 2015-07-22 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.545 Å) | | Cite: | Transthyretin Binding Heterogeneity and Anti-amyloidogenic Activity of Natural Polyphenols and Their Metabolites.
J.Biol.Chem., 290, 2015
|
|
5CRB
 
 | | Crystal Structure of SdeA DUB | | Descriptor: | SdeA | | Authors: | Sheedlo, M.J, Qiu, J, Luo, Z.Q, Das, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1HQG
 
 | | CRYSTAL STRUCTURE OF THE H141C ARGINASE VARIANT COMPLEXED WITH PRODUCTS ORNITHINE AND UREA | | Descriptor: | ARGINASE 1, L-ornithine, MANGANESE (II) ION, ... | | Authors: | Cox, J.D, Cama, E, Colleluori, D.M, Ash, D.E, Christianson, D.W. | | Deposit date: | 2000-12-16 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic and metabolic inferences from the binding of substrate analogues and products to arginase.
Biochemistry, 40, 2001
|
|
6HYP
 
 | | Rea1 Wild type ADP state (AAA+ ring part) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Midasin,Midasin | | Authors: | Sosnowski, P, Urnavicius, L, Boland, A, Fagiewicz, R, Busselez, J, Papai, G, Schmidt, H. | | Deposit date: | 2018-10-22 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The CryoEM structure of the Saccharomyces cerevisiae ribosome maturation factor Rea1.
Elife, 7, 2018
|
|
1HWY
 
 | | BOVINE GLUTAMATE DEHYDROGENASE COMPLEXED WITH NAD AND 2-OXOGLUTARATE | | Descriptor: | 2-OXOGLUTARIC ACID, GLUTAMATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Smith, T.J, Peterson, P.E, Schmidt, T, Fang, J, Stanley, C.A. | | Deposit date: | 2001-01-10 | | Release date: | 2001-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of bovine glutamate dehydrogenase complexes elucidate the mechanism of purine regulation.
J.Mol.Biol., 307, 2001
|
|
1ZM9
 
 | | Structure of eEF2-ETA in complex with PJ34 | | Descriptor: | Elongation factor 2, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, exotoxin A | | Authors: | Joergensen, R, Merrill, A.R, Yates, S.P, Marquez, V.E, Schwan, A.L, Boesen, T, Andersen, G.R. | | Deposit date: | 2005-05-10 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry.
Nature, 436, 2005
|
|
4NM3
 
 | | Crystal structure of GSK-3/Axin complex bound to phosphorylated N-terminal auto-inhibitory pS9 peptide | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADENOSINE-5'-DIPHOSPHATE, Axin-1, ... | | Authors: | Chu, M.L.-H, Stamos, J.L, Enos, M.D, Shah, N, Weis, W.I. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of GSK-3 inhibition by N-terminal phosphorylation and by the Wnt receptor LRP6.
Elife, 3, 2014
|
|
5YYL
 
 | | Structure of Major Royal Jelly Protein 1 Oligomer | | Descriptor: | (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tian, W, Chen, Z. | | Deposit date: | 2017-12-10 | | Release date: | 2018-08-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Architecture of the native major royal jelly protein 1 oligomer.
Nat Commun, 9, 2018
|
|
1EH4
 
 | | BINARY COMPLEX OF CASEIN KINASE-1 FROM S. POMBE WITH AN ATP COMPETITIVE INHIBITOR, IC261 | | Descriptor: | 3-[(2,4,6-TRIMETHOXY-PHENYL)-METHYLENE]-INDOLIN-2-ONE, CASEIN KINASE-1, SULFATE ION | | Authors: | Mashhoon, N, Demaggio, A.J, Tereshko, V, Bergmeier, S.C, Egli, M, Hoekstra, M.F, Kuret, J. | | Deposit date: | 2000-02-18 | | Release date: | 2001-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Conformation-Selective Casein Kinase-1 Inhibitor
J.Biol.Chem., 275, 2000
|
|
3BG0
 
 | | Architecture of a Coat for the Nuclear Pore Membrane | | Descriptor: | Nucleoporin NUP145, Protein SEC13 homolog | | Authors: | Hoelz, A. | | Deposit date: | 2007-11-23 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Architecture of a coat for the nuclear pore membrane.
Cell(Cambridge,Mass.), 131, 2007
|
|
5CXA
 
 | | Crystal structure of the catalytic domain of Human MMP12 in complex with a carboxylate inhibitor related to RXP470 | | Descriptor: | (R)-4-(((S)-1-(((S)-1-amino-4-carboxy-1-oxobutan-2-yl)amino)-4-carboxy-1-oxobutan-2-yl)amino)-3-((3-(3'-chloro-[1,1'-biphenyl]-4-yl)isoxazol-5-yl)methyl)-4-oxobutanoate, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Rouanet-Mehouas, C, Devel, L, Dive, V, Stura, E.A. | | Deposit date: | 2015-07-28 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Zinc-Metalloproteinase Inhibitors: Evaluation of the Complex Role Played by the Zinc-Binding Group on Potency and Selectivity.
J. Med. Chem., 60, 2017
|
|
4NMR
 
 | |
