9H28
 
 | | Alternative conformation LGTV with TBEV prME | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, ... | | Authors: | Bisikalo, K, Rosendal, E. | | Deposit date: | 2024-10-11 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The influence of the pre-membrane and envelope proteins on structure, pathogenicity and tropism of tick-borne encephalitis virus
Biorxiv, 2024
|
|
7B5G
 
 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS3(Nb14527) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, Nanobody Nb14527, ... | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2020-12-03 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
7R0D
 
 | |
8F4Y
 
 | | Crystal Structure of SARS-CoV-2 2'-O-Methyltransferase in Complex with Compound 5a covalently bound to nsp16 and nsp10 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, 4-[2-(2,4-dichlorophenyl)ethyl]-6-(trifluoromethyl)pyrimidin-2-ol, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8F4S
 
 | | Crystal Structure of the SARS-CoV-2 2'-O-Methyltransferase with Compound 5a bound to the Cryptic Pocket of nsp16 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8PNT
 
 | | Structure of the human nuclear cap-binding complex bound to PHAX and m7G-capped RNA | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Nuclear cap-binding protein subunit 1, Nuclear cap-binding protein subunit 2, ... | | Authors: | Dubiez, E, Pellegrini, E, Foucher, A.E, Cusack, S, Kadlec, J. | | Deposit date: | 2023-07-01 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis for competitive binding of productive and degradative co-transcriptional effectors to the nuclear cap-binding complex.
Cell Rep, 43, 2024
|
|
4KGZ
 
 | | The R state structure of E. coli ATCase with UTP and Magnesium bound | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, MAGNESIUM ION, ... | | Authors: | Cockrell, G.M, Zheng, Y, Guo, W, Peterson, A.W, Kantrowitz, E.R. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New Paradigm for Allosteric Regulation of Escherichia coli Aspartate Transcarbamoylase.
Biochemistry, 52, 2013
|
|
7PUK
 
 | | Crystal structure of Endoglycosidase E GH18 domain from Enterococcus faecalis in complex with Man5 product | | Descriptor: | Beta-N-acetylhexosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Garcia-Alija, M, Du, J.J, Trastoy, B, Sundberg, E.J, Guerin, M. | | Deposit date: | 2021-09-30 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Mechanism of cooperative N-glycan processing by the multi-modular endoglycosidase EndoE.
Nat Commun, 13, 2022
|
|
7S87
 
 | | Crystal Structure of Dihydroorotate dehydrogenase from Plasmodium falciparum in complex with Orotate, FMN, and inhibitor NCGC00600348-01 | | Descriptor: | (4R)-3-methyl-5-[(4R)-4-methyl-3,4-dihydroisoquinolin-2(1H)-yl]thieno[2,3-e][1,2,4]triazolo[4,3-c]pyrimidine, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-09-17 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Dihydroorotate dehydrogenase from Plasmodium falciparum in complex with Orotate, FMN, and inhibitor NCGC00600348-01
To Be Published
|
|
9BE5
 
 | | Cryo-EM structure of Human Nucleosome collected by EPU on Glacios at 3.3 Angstrom resolution | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2A type 1-B/E, ... | | Authors: | Jia, L, Ruben, E.A, Olsen, S.K, Wasmuth, E.V. | | Deposit date: | 2024-04-14 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of Human Nucleosome collected by EPU on Glacios at 3.3 Angstrom resolution
To Be Published
|
|
4KGV
 
 | | The R state structure of E. coli ATCase with ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, ... | | Authors: | Cockrell, G.M, Zheng, Y, Guo, W, Peterson, A.W, Kantrowitz, E.R. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New Paradigm for Allosteric Regulation of Escherichia coli Aspartate Transcarbamoylase.
Biochemistry, 52, 2013
|
|
8PMX
 
 | | rat HEV P domain in complex with glycan-sensitive nAb p60.12 | | Descriptor: | Fab p60.12-HC, Fab p60.12-LC, Pro-secreted protein ORF2 | | Authors: | Ssebyatika, G, Krey, T. | | Deposit date: | 2023-06-29 | | Release date: | 2024-07-10 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (3.922 Å) | | Cite: | Broadly neutralizing antibodies isolated from HEV convalescents confer protective effects in human liver-chimeric mice.
Nat Commun, 16, 2025
|
|
6M4I
 
 | |
4KGX
 
 | | The R state structure of E. coli ATCase with CTP bound | | Descriptor: | Aspartate carbamoyltransferase, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Cockrell, G.M, Zheng, Y, Guo, W, Peterson, A.W, Kantrowitz, E.R. | | Deposit date: | 2013-04-29 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | New Paradigm for Allosteric Regulation of Escherichia coli Aspartate Transcarbamoylase.
Biochemistry, 52, 2013
|
|
6MID
 
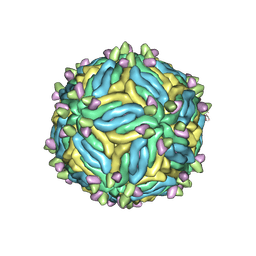 | |
8B5Q
 
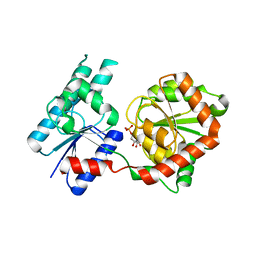 | |
8B62
 
 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-galactose | | Descriptor: | GALACTOSE-URIDINE-5'-DIPHOSPHATE, GLYCEROL, UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8B63
 
 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-GalNAc | | Descriptor: | ACETATE ION, UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-25 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
8B5S
 
 | | Crystal Structure of P. aeruginosa WaaG in complex with UDP-glucose | | Descriptor: | UDP-glucose:(Heptosyl) LPS alpha 1,3-glucosyltransferase WaaG, URIDINE-5'-DIPHOSPHATE, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Scaletti, E, Gustafsson Westergren, R, Stenmark, P. | | Deposit date: | 2022-09-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and functional insights into the Pseudomonas aeruginosa glycosyltransferase WaaG and the implications for lipopolysaccharide biosynthesis.
J.Biol.Chem., 299, 2023
|
|
6ZOF
 
 | | Fusidic acid binding to the TM7/TM8 groove of AcrB-F380A T protomer | | Descriptor: | DARPIN, DECANE, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Tam, H.K, Foong, W.E, Pos, K.M. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Allosteric drug transport mechanism of multidrug transporter AcrB.
Nat Commun, 12, 2021
|
|
7QGO
 
 | | Human CD73 (ecto 5'-nucleotidase) in complex with MRS4602 (a 3-methyl-CMPCP derivative, compound 21 in paper) in the closed state (crystal form III) | | Descriptor: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-[(4~{E})-4-[(4-methoxycarbonylphenyl)methoxyimino]-3-methyl-2-oxidanylidene-pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | Authors: | Strater, N. | | Deposit date: | 2021-12-09 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure-Activity Relationship of 3-Methylcytidine-5'-alpha , beta-methylenediphosphates as CD73 Inhibitors.
J.Med.Chem., 65, 2022
|
|
8BT7
 
 | | Notum Inhibitor ARUK3004903 | | Descriptor: | 1,2-ETHANEDIOL, 1-[3,4-bis(chloranyl)-5-methyl-indol-1-yl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BT5
 
 | | Notum Inhibitor ARUK3004877 | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-fluoranylspiro[2~{H}-indole-3,1'-cyclobutane]-1-yl)ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-27 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BSQ
 
 | | Notum Inhibitor ARUK3006561 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8BTC
 
 | | Notum Inhibitor ARUK3004558 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4,5-bis(chloranyl)-2,3-dihydroindol-1-yl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
