5FYL
 
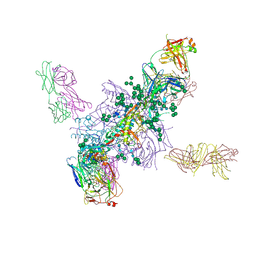 | | Crystal Structure at 3.7 A Resolution of Fully Glycosylated HIV-1 Clade A BG505 SOSIP.664 Prefusion Env Trimer in Complex with Broadly Neutralizing Antibodies PGT122 and 35O22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 ANTIBODY FAB HEAVY CHAIN, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Thomas, P.V, Kwong, P.D. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B and G
Cell(Cambridge,Mass.), 165, 2016
|
|
5ELY
 
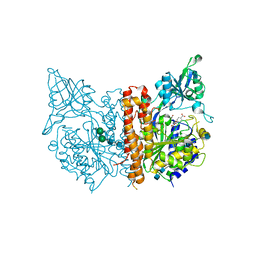 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a hydroxamate inhibitor JHU242 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(2~{R})-2-carboxy-5-(oxidanylamino)-5-oxidanylidene-pentyl]benzoic acid, ... | | Authors: | Barinka, C, Novakova, Z, Pavlicek, J. | | Deposit date: | 2015-11-05 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Unprecedented Binding Mode of Hydroxamate-Based Inhibitors of Glutamate Carboxypeptidase II: Structural Characterization and Biological Activity.
J.Med.Chem., 59, 2016
|
|
7US6
 
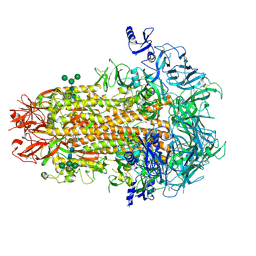 | | Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the proximal conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7LKP
 
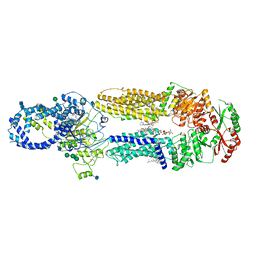 | | Structure of ATP-free human ABCA4 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, F, Lee, J, Chen, J. | | Deposit date: | 2021-02-02 | | Release date: | 2021-03-03 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Molecular structures of the eukaryotic retinal importer ABCA4.
Elife, 10, 2021
|
|
4YYA
 
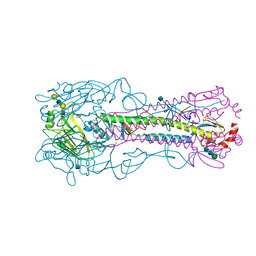 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) in complex with avian receptor analog 3'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
5FUK
 
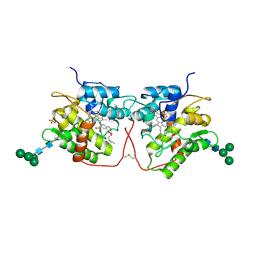 | |
7USA
 
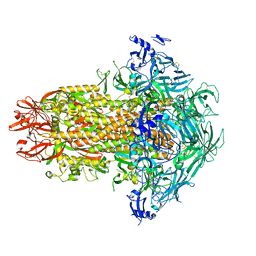 | | Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7LKZ
 
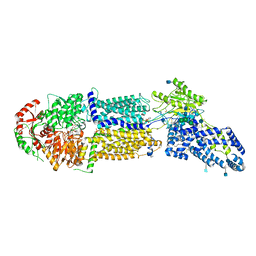 | | Structure of ATP-bound human ABCA4 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Liu, F, Lee, J, Chen, J. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Molecular structures of the eukaryotic retinal importer ABCA4.
Elife, 10, 2021
|
|
1EOM
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF ENDO-BETA-N-ACETYLGLUCOSAMINIDASE F3 WITH A BIANTENNARY COMPLEX OCTASACCHARIDE | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE F3, SULFATE ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Waddling, C.A, Plummer Jr, T.H, Tarentino, A.L, Van Roey, P. | | Deposit date: | 2000-03-23 | | Release date: | 2000-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the substrate specificity of endo-beta-N-acetylglucosaminidase F(3).
Biochemistry, 39, 2000
|
|
6H41
 
 | |
5FC6
 
 | | Murine SMPDL3A in complex with ADP analog AMPCP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Gorelik, A, Illes, K, Superti-Furga, G, Nagar, B. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Structural Basis for Nucleotide Hydrolysis by the Acid Sphingomyelinase-like Phosphodiesterase SMPDL3A.
J.Biol.Chem., 291, 2016
|
|
7P77
 
 | | SARS-CoV-2 spike protein in complex with sybody#15 and sybody#68 in a 3up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P7A
 
 | | SARS-CoV-2 spike protein in complex with sybody#68 in a 2up/1flexible conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (4.76 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
6XU0
 
 | |
7P78
 
 | | SARS-CoV-2 spike protein in complex with sybody#15 and sybody#68 in a 1up/1up-out/1down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
7P79
 
 | | SARS-CoV-2 spike protein in complex with sybodyb#15 in a 1up/1up-out/1down conformation. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(5-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Walter, J.D, Hutter, C.A.J, Garaeva, A.A, Scherer, M, Zimmermann, I, Wyss, M, Rheinberger, J, Ruedin, Y, Earp, J.C, Egloff, P, Sorgenfrei, M, Huerlimann, L.M, Gonda, I, Meier, G, Remm, S, Thavarasah, S, Zimmer, G, Slotboom, D.J, Paulino, C, Plattet, P, Seeger, M.A. | | Deposit date: | 2021-07-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Biparatopic sybodies neutralize SARS-CoV-2 variants of concern and mitigate drug resistance.
Embo Rep., 23, 2022
|
|
4YYB
 
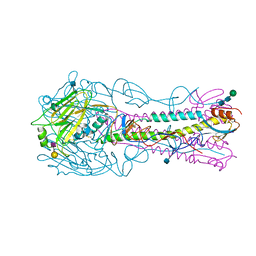 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) in complex with human receptor analog 6'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) at 2.6 Angstroms resolution
To Be Published
|
|
1E6S
 
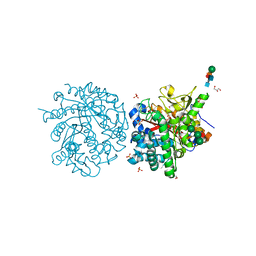 | | MYROSINASE FROM SINAPIS ALBA with bound gluco-hydroximolactam and sulfate | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Burmeister, W.P. | | Deposit date: | 2000-08-23 | | Release date: | 2000-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High Resolution X-Ray Crystallography Shows that Ascorbate is a Cofactor for Myrosinase and Substitutes for the Function of the Catalytic Base
J.Biol.Chem., 275, 2000
|
|
5J4B
 
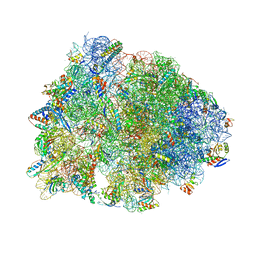 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with cisplatin (co-crystallized) and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Melnikov, S.V, Soll, D, Steitz, T.A, Polikanov, Y.S. | | Deposit date: | 2016-03-31 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into RNA binding by the anticancer drug cisplatin from the crystal structure of cisplatin-modified ribosome.
Nucleic Acids Res., 44, 2016
|
|
6O9E
 
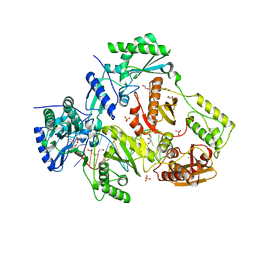 | | Structure of HIV-1 Reverse Transcriptase in complex with DNA and INDOPY-1 | | Descriptor: | 5-methyl-1-(4-nitrophenyl)-2-oxo-2,5-dihydro-1H-pyrido[3,2-b]indole-3-carbonitrile, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ruiz, F.X, Hoang, A, Das, K, Arnold, E. | | Deposit date: | 2019-03-13 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of HIV-1 Inhibition by Nucleotide-Competing Reverse Transcriptase Inhibitor INDOPY-1.
J.Med.Chem., 62, 2019
|
|
1ECE
 
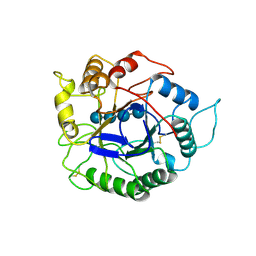 | | ACIDOTHERMUS CELLULOLYTICUS ENDOCELLULASE E1 CATALYTIC DOMAIN IN COMPLEX WITH A CELLOTETRAOSE | | Descriptor: | ENDOCELLULASE E1, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Sakon, J, Thomas, S.R, Himmel, M.E, Karplus, P.A. | | Deposit date: | 1996-04-04 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of thermostable family 5 endocellulase E1 from Acidothermus cellulolyticus in complex with cellotetraose.
Biochemistry, 35, 1996
|
|
8D43
 
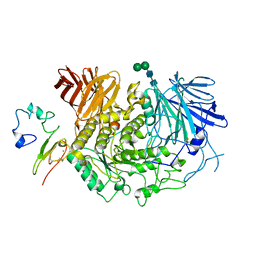 | |
5LVX
 
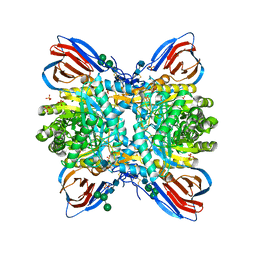 | | Crystal structure of glucocerebrosidase with an inhibitory quinazoline modulator | | Descriptor: | 11-[(2~{R})-2-[(2-pyridin-3-ylquinazolin-4-yl)amino]-2,3-dihydro-1~{H}-inden-5-yl]undec-10-ynoic acid, 11-[(2~{S})-2-[(2-pyridin-3-ylquinazolin-4-yl)amino]-2,3-dihydro-1~{H}-inden-5-yl]undec-10-ynoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, J, Chen, L, Skinner, O.S, Lansbury, P, Skerlj, R, Mrosek, M, Heunisch, U, Krapp, S, Weigand, S, Charrow, J, Schwake, M, Kelleher, N.L, Silverman, R.B, Krainc, D. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | beta-Glucocerebrosidase Modulators Promote Dimerization of beta-Glucocerebrosidase and Reveal an Allosteric Binding Site.
J. Am. Chem. Soc., 140, 2018
|
|
6FU7
 
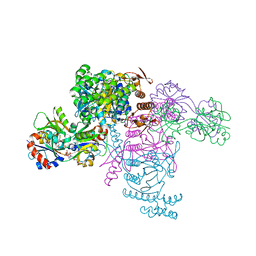 | | ATP phosphoribosyltransferase (HisZG ATPPRT) from Psychrobacter arcticus in complex with PRATP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP phosphoribosyltransferase, ATP phosphoribosyltransferase regulatory subunit, ... | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2018-02-26 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
3EYZ
 
 | | Cocrystal structure of Bacillus fragment DNA polymerase I with duplex DNA (open form) | | Descriptor: | 5'-D(*DAP*DTP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DAP*DGP*DGP*DA)-3', 5'-D(*DCP*DCP*DTP*DGP*DAP*DCP*DTP*DCP*DGP*DC)-3', DNA polymerase I, ... | | Authors: | Warren, J.J, Wu, E.Y, Golosov, A.A, Karplus, M, Beese, L.S. | | Deposit date: | 2008-10-22 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Mechanism of the Translocation Step in DNA Replication by DNA Polymerase I: A Computer Simulation Analysis.
Structure, 18, 2010
|
|
