6EO9
 
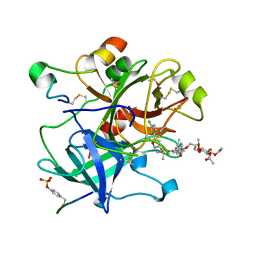 | | Crystal structure of thrombin in complex with a novel glucose-conjugated potent inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, Hirudin variant-2, N-(2-{[5-(5-chlorothiophen-2-yl)-1,2-oxazol-3-yl]methoxy}-6-{3-[(2,3,4,6-tetra-O-acetyl-beta-D-glucopyranosyl)oxy]propoxy}phenyl)-1-(propan-2-yl)piperidine-4-carboxamide, ... | | Authors: | Belviso, B.D, Caliandro, R, Aresta, B.M, De Candia, M, Altomare, C.D. | | Deposit date: | 2017-10-09 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How a beta-D-glucoside side chain enhances binding affinity to thrombin of inhibitors bearing 2-chlorothiophene as P1 moiety: crystallography, fragment deconstruction study, and evaluation of antithrombotic properties.
J. Med. Chem., 57, 2014
|
|
1TP8
 
 | | CRYSTAL STRUCTURE OF A GALACTOSE SPECIFIC LECTIN FROM ARTOCARPUS HIRSUTA IN COMPLEX WITH METHYL-a-D-GALACTOSE | | Descriptor: | AGGLUTININ ALPHA CHAIN, AGGLUTININ BETA CHAIN, methyl alpha-D-galactopyranoside | | Authors: | Rao, K.N, Suresh, C.G, Katre, U.V, Gaikwad, S.M, Khan, M.I. | | Deposit date: | 2004-06-16 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two orthorhombic crystal structures of a galactose-specific lectin from Artocarpus hirsuta in complex with methyl-alpha-D-galactose.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4UAG
 
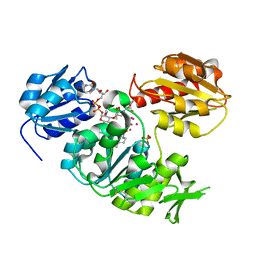 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | SULFATE ION, UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, UNKNOWN ATOM OR ION, ... | | Authors: | Bertrand, J, Auger, G, Martin, L, Fanchon, E, Blanot, D, Le Beller, D, Van Heijenoort, J, Dideberg, O. | | Deposit date: | 1999-03-09 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Determination of the MurD mechanism through crystallographic analysis of enzyme complexes.
J.Mol.Biol., 289, 1999
|
|
9XIA
 
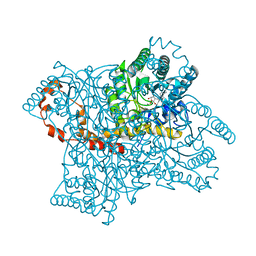 | |
8BZI
 
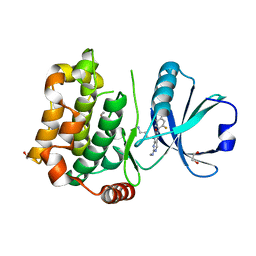 | | Human MST3 (STK24) kinase in complex with inhibitor MR39 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2,5-bis(fluoranyl)-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Tesch, R, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Shifting the selectivity of pyrido[2,3-d]pyrimidin-7(8H)-one inhibitors towards the salt-inducible kinase (SIK) subfamily.
Eur.J.Med.Chem., 254, 2023
|
|
8BSA
 
 | | Vc1313-LBD bound to D-arginine | | Descriptor: | D-ARGININE, Methyl-accepting chemotaxis protein | | Authors: | ter Beek, J, Berntsson, R.P.-A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D-amino acids signal a stress-dependent run-away response in Vibrio cholerae.
Nat Microbiol, 8, 2023
|
|
8BSB
 
 | | Vc1313-LBD bound to D-lysine | | Descriptor: | D-LYSINE, Methyl-accepting chemotaxis protein | | Authors: | ter Beek, J, Berntsson, R.P.-A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D-amino acids signal a stress-dependent run-away response in Vibrio cholerae.
Nat Microbiol, 8, 2023
|
|
6GVA
 
 | | CDK2/cyclin A2 in complex with pyrazolo[4,3-d]pyrimidine inhibitor LGR4455 | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-azanylethylsulfanyl)-3-propan-2-yl-~{N}-[(4-pyridin-2-ylphenyl)methyl]-2~{H}-pyrazolo[4,3-d]pyrimidin-7-amine, BROMIDE ION, ... | | Authors: | Skerlova, J, Rezacova, P. | | Deposit date: | 2018-06-20 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3,5,7-Substituted Pyrazolo[4,3- d]pyrimidine Inhibitors of Cyclin-Dependent Kinases and Their Evaluation in Lymphoma Models.
J.Med.Chem., 62, 2019
|
|
4X56
 
 | | Structure of the class D Beta-Lactamase OXA-160 V130D in Acyl-Enzyme Complex with Ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Class D beta-lactamase OXA-160 | | Authors: | Clasman, J.R, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D beta-Lactamases from Acinetobacter baumannii.
Biochemistry, 54, 2015
|
|
186D
 
 | |
7RN3
 
 | | hyen D solution structure | | Descriptor: | Cyclotide hyen-D | | Authors: | Du, Q, Huang, Y.H, Craik, D.J, Wang, C.K. | | Deposit date: | 2021-07-29 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Mutagenesis of bracelet cyclotide hyen D reveals functionally and structurally critical residues for membrane binding and cytotoxicity.
J.Biol.Chem., 298, 2022
|
|
4X53
 
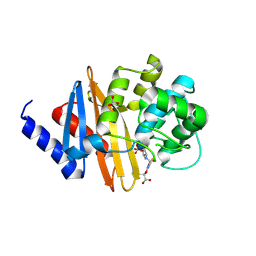 | | Structure of the class D Beta-Lactamase OXA-160 V130D in Acyl-Enzyme Complex with Aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, BICARBONATE ION, Class D beta-lactamase OXA-160 | | Authors: | Clasman, J.R, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D beta-Lactamases from Acinetobacter baumannii.
Biochemistry, 54, 2015
|
|
7AD3
 
 | | Class D GPCR Ste2 dimer coupled to two G proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-factor mating pheromone, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Velazhahan, V, Tate, C. | | Deposit date: | 2020-09-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the class D GPCR Ste2 dimer coupled to two G proteins.
Nature, 589, 2021
|
|
6OJH
 
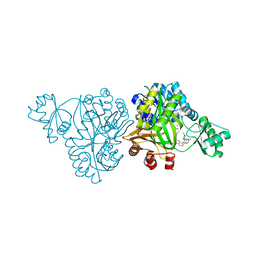 | | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with (R)-7-(3-aminopyrrolidin-1-yl)-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 7-[(3R)-3-aminopyrrolidin-1-yl]-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine, ACETATE ION, Biotin carboxylase, ... | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-11 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with (R)-7-(3-aminopyrrolidin-1-yl)-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine
To Be Published
|
|
7RKE
 
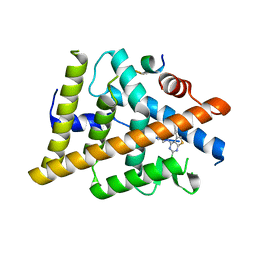 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S in Complex with 4-(((2-chloro-5-phenylthieno[2,3-d]pyrimidin-4-yl)amino)methyl)phenol and GRIP Peptide | | Descriptor: | 4-{[(2-chloro-5-phenylthieno[2,3-d]pyrimidin-4-yl)amino]methyl}phenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Joiner, C, Sammeta, V.K.R, Norris, J.D, McDonnell, D.P, Willson, T.M, Fanning, S.W. | | Deposit date: | 2021-07-22 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A New Chemotype of Chemically Tractable Nonsteroidal Estrogens Based on a Thieno[2,3- d ]pyrimidine Core.
Acs Med.Chem.Lett., 13, 2022
|
|
4XTU
 
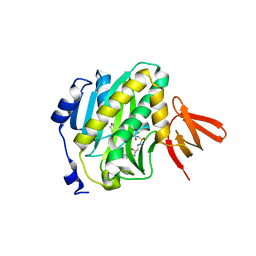 | | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor (N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide) | | Descriptor: | Bifunctional ligase/repressor BirA, N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide | | Authors: | De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2015-01-24 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6500299 Å) | | Cite: | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor (N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide)
To be Published
|
|
8R1X
 
 | |
7RNM
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with 2-(2-Chloro-5-phenylthieno[2,3-d]pyrimidin-4-yl)isoindolin-5-ol and GRIP Peptide | | Descriptor: | 2-(2-chloro-5-phenylthieno[2,3-d]pyrimidin-4-yl)-2,3-dihydro-1H-isoindol-5-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Joiner, C, Sammeta, V.K.R, Norris, J.D, McDonnell, D.P, Willson, T.M, Fanning, S.W. | | Deposit date: | 2021-07-29 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Determinants of the Binding and Activation of Estrogen Receptor alpha by Phenolic Thieno[2,3-d]pyrimidines
Helv.Chim.Acta, 106, 2023
|
|
2WI7
 
 | | Orally Active 2-Amino Thienopyrimidine Inhibitors of the Hsp90 Chaperone | | Descriptor: | 2-amino-4-[2,4-dichloro-5-(2-pyrrolidin-1-ylethoxy)phenyl]-N-ethylthieno[2,3-d]pyrimidine-6-carboxamide, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Brough, P.A, Barril, X, Borgognoni, J, Chene, P, Davies, N.G.M, Davis, B, Drysdale, M.J, Dymock, B, Eccles, S.A, Garcia-Echeverria, C, Fromont, C, Hayes, A, Hubbard, R.E, Jordan, A.M, Rugaard-Jensen, M, Massey, A, Merret, A, Padfield, A, Parsons, R, Radimerski, T, Raynaud, F.I, Robertson, A, Roughley, S.D, Schoepfer, J, Simmonite, H, Surgenor, A, Valenti, M, Walls, S, Webb, P, Wood, M, Workman, P, Wright, L.M. | | Deposit date: | 2009-05-08 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Combining Hit Identification Strategies: Fragment- Based and in Silico Approaches to Orally Active 2-Aminothieno[2,3-D]Pyrimidine Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 52, 2009
|
|
6PSA
 
 | | PIE12 D-PEPTIDE AGAINST HIV ENTRY (IN COMPLEX WITH IQN17 Q577R RESISTANCE MUTANT) | | Descriptor: | CHLORIDE ION, IQN17, PIE12 D-peptide | | Authors: | Hill, C.P, Whitby, F.G, Kay, M, Weinstock, M. | | Deposit date: | 2019-07-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of resistance to a potent D-peptide HIV entry inhibitor.
Retrovirology, 16, 2019
|
|
9HTA
 
 | | Dispersin from Terribacillus saccharophilus Dispts2 in complex with NAG-thiazoline | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Dispersin | | Authors: | Males, A, Moroz, O.V, Blagova, E, Munch, A, Hansen, G.H, Johansen, A.H, Ostergaard, L.H, Segura, D.R, Eddenden, A, Due, A.V, Gudmand, M, Salomon, J, Sorensen, S.R, Franco Cairo, J.P.L, Pache, R.A, Vejborg, R.M, Bhosale, S, Vocadlo, D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2024-12-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Expansion of the diversity of dispersin scaffolds.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
2WI6
 
 | | Orally Active 2-Amino Thienopyrimidine Inhibitors of the Hsp90 Chaperone | | Descriptor: | 2-AMINO-4-(2,4-DICHLOROPHENYL)-N-ETHYLTHIENO[2,3-D]PYRIMIDINE-6-CARBOXAMIDE, HEAT SHOCK PROTEIN, HSP90-ALPHA | | Authors: | Brough, P.A, Barril, X, Borgognoni, J, Chene, P, Davies, N.G.M, Davis, B, Drysdale, M.J, Dymock, B, Eccles, S.A, Garcia-Echeverria, C, Fromont, C, Hayes, A, Hubbard, R.E, Jordan, A.M, Rugaard-Jensen, M, Massey, A, Merret, A, Padfield, A, Parsons, R, Radimerski, T, Raynaud, F.I, Robertson, A, Roughley, S.D, Schoepfer, J, Simmonite, H, Surgenor, A, Valenti, M, Walls, S, Webb, P, Wood, M, Workman, P, Wright, L.M. | | Deposit date: | 2009-05-08 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Combining Hit Identification Strategies: Fragment- Based and in Silico Approaches to Orally Active 2-Aminothieno[2,3-D]Pyrimidine Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 52, 2009
|
|
8XIV
 
 | | Production of D-psicose | | Descriptor: | COBALT (II) ION, D-fructose, D-tagatose 3-epimerase, ... | | Authors: | Guo, D. | | Deposit date: | 2023-12-20 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Production of D-psicose
To Be Published
|
|
8P3A
 
 | | bacteriophage T5 l-alanoyl-d-glutamate peptidase Zn2+/Ca2+ form | | Descriptor: | CALCIUM ION, L-alanyl-D-glutamate peptidase, ZINC ION | | Authors: | Prokhorov, D.A, Kutyshenko, V.P, Mikoulinskaia, G.V. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-05 | | Last modified: | 2025-01-15 | | Method: | SOLUTION NMR | | Cite: | Conservative Tryptophan Residue in the Vicinity of an Active Site of the M15 Family l,d-Peptidases: A Key Element in the Catalysis.
Int J Mol Sci, 24, 2023
|
|
4WWH
 
 | | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEG_1704, TARGET EFI-510967) WITH BOUND D-GALACTOSE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, beta-D-galactopyranose | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ABC TRANSPORTER SOLUTE BINDING PROTEIN (IPR025997) FROM MYCOBACTERIUM SMEGMATIS (MSMEG_1704, TARGET EFI-510967) WITH BOUND D-GALACTOSE
To be published
|
|
