8DPX
 
 | | Preligand association structure of DR5 | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Du, G, Zhao, L, Chou, J.J. | | Deposit date: | 2022-07-17 | | Release date: | 2023-02-15 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory structure of preligand association state implicates a new strategy to attain effective DR5 receptor activation.
Cell Res., 33, 2023
|
|
6H67
 
 | | Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Sanz-Murillo, M, Xu, J, Gil-Carton, D, Wang, D, Fernandez-Tornero, C. | | Deposit date: | 2018-07-26 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of RNA polymerase I stalling at UV light-induced DNA damage.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6H68
 
 | | Yeast RNA polymerase I elongation complex stalled by cyclobutane pyrimidine dimer (CPD) with fully-ordered A49 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Sanz-Murillo, M, Xu, J, Gil-Carton, D, Wang, D, Fernandez-Tornero, C. | | Deposit date: | 2018-07-26 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of RNA polymerase I stalling at UV light-induced DNA damage.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6MF0
 
 | | Crystal Structure Determination of Human/Porcine Chimera Coagulation Factor VIII | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Smith, I.W, Spiegel, P.C. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.19999886 Å) | | Cite: | The 3.2 angstrom structure of a bioengineered variant of blood coagulation factor VIII indicates two conformations of the C2 domain.
J.Thromb.Haemost., 18, 2020
|
|
6JSH
 
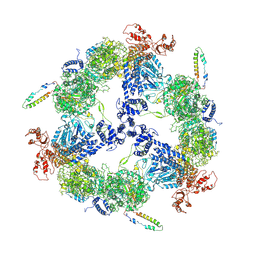 | | Apo-state Fatty Acid Synthase | | Descriptor: | Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Qiu, S.W, Liu, S. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Modulation of fatty acid synthase by ATR checkpoint kinase Rad3.
J Mol Cell Biol, 11, 2019
|
|
6HKO
 
 | | Yeast RNA polymerase I elongation complex bound to nucleotide analog GMPCPP | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-07 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
6HA1
 
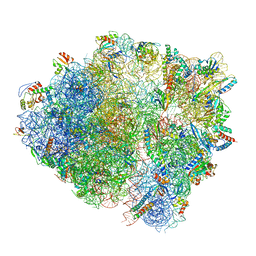 | | Cryo-EM structure of a 70S Bacillus subtilis ribosome translating the ErmD leader peptide in complex with telithromycin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Graf, M, Huter, P, Abdelshahid, M, Novacek, J, Wilson, D.N. | | Deposit date: | 2018-08-07 | | Release date: | 2018-08-29 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for antibiotic resistance mediated by theBacillus subtilisABCF ATPase VmlR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4YM7
 
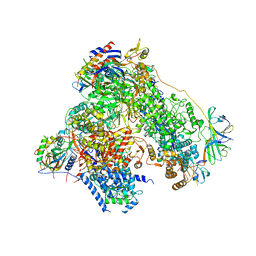 | | RNA polymerase I structure with an alternative dimer hinge | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Kostrewa, D, Kuhn, C.-D, Engel, C, Cramer, P. | | Deposit date: | 2015-03-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | An alternative RNA polymerase I structure reveals a dimer hinge.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6HLQ
 
 | | Yeast RNA polymerase I* elongation complex bound to nucleotide analog GMPCPP | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-11 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
6JSI
 
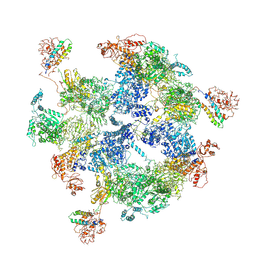 | | Co-purified Fatty Acid Synthase | | Descriptor: | Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Qiu, S.W, Liu, S. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Modulation of fatty acid synthase by ATR checkpoint kinase Rad3.
J Mol Cell Biol, 11, 2019
|
|
6HA8
 
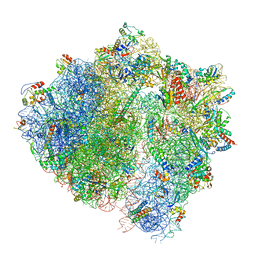 | | Cryo-EM structure of the ABCF protein VmlR bound to the Bacillus subtilis ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Graf, M, Huter, P, Abdelshahid, M, Novacek, J, Wilson, D.N. | | Deposit date: | 2018-08-07 | | Release date: | 2018-08-29 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for antibiotic resistance mediated by theBacillus subtilisABCF ATPase VmlR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZX3
 
 | |
5ZX2
 
 | |
6NMI
 
 | | Cryo-EM structure of the human TFIIH core complex | | Descriptor: | CDK-activating kinase assembly factor MAT1, General transcription and DNA repair factor IIH helicase subunit XPB, General transcription and DNA repair factor IIH helicase subunit XPD, ... | | Authors: | Greber, B.J, Toso, D, Fang, J, Nogales, E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-03-13 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The complete structure of the human TFIIH core complex.
Elife, 8, 2019
|
|
5ENB
 
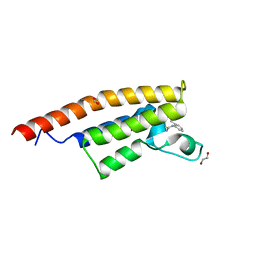 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with o-Tolylthiourea (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-methylphenyl)thiourea, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENI
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with compound-13 N11537 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | PH-interacting protein, ~{N}-[[2,6-bis(chloranyl)phenyl]methyl]-2-oxidanyl-ethanamide | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENE
 
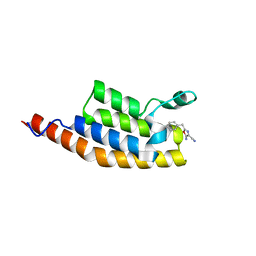 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with 5-Amino-2-benzyl-1,3-oxazole-4-carbonitrile (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 5-azanyl-2-(phenylmethyl)-1,3-oxazole-4-carbonitrile, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Amin, J, Szykowska, A, Burgess-Brown, N, Spencer, J, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENJ
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with compound-14 N11530 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | MAGNESIUM ION, PH-interacting protein, ~{N}-[[2,6-bis(chloranyl)phenyl]methyl]-~{N}-methyl-ethanamide | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENC
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with N-(2,6-Dichlorobenzyl)acetamide (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, PH-interacting protein, ~{N}-[[2,6-bis(chloranyl)phenyl]methyl]ethanamide | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5FCJ
 
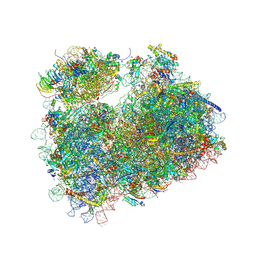 | | Structure of the anisomycin-containing uL3 W255C mutant 80S yeast ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Mailliot, J, Garreau de Loubresse, N, Yusupova, G, Dinman, J.D, Yusupov, M. | | Deposit date: | 2015-12-15 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of the uL3 Mutant Ribosome: Illustration of the Importance of Ribosomal Proteins for Translation Efficiency.
J.Mol.Biol., 428, 2016
|
|
5FCI
 
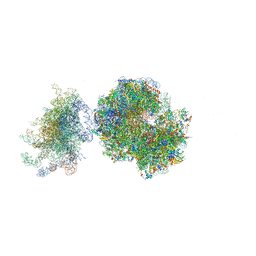 | | Structure of the vacant uL3 W255C mutant 80S yeast ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Mailliot, J, Garreau de Loubresse, N, Yusupova, G, Dinman, J.D, Yusupov, M. | | Deposit date: | 2015-12-15 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structures of the uL3 Mutant Ribosome: Illustration of the Importance of Ribosomal Proteins for Translation Efficiency.
J.Mol.Biol., 428, 2016
|
|
5ENF
 
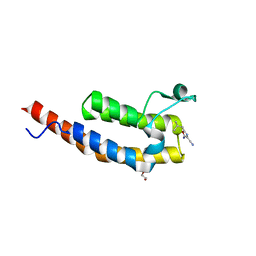 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with fragment-4 N10142 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-2-(2-methylpropyl)-1,3-oxazole-4-carbonitrile, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Amin, J, Szykowska, A, Burgess-Brown, N, Spencer, J, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5ENH
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with compound-12 N11528 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | PH-interacting protein, ~{N}-[(2,6-dimethoxyphenyl)methyl]ethanamide | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
5G5L
 
 | | RNA polymerase I-Rrn3 complex at 4.8 A resolution | | Descriptor: | DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA14, ... | | Authors: | Engel, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-27 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | RNA Polymerase I-Rrn3 Complex at 4.8 A Resolution
Nat.Commun., 7, 2016
|
|
8G6I
 
 | | Coagulation factor VIII bound to a patient-derived anti-C1 domain antibody inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor VIII chimera from human and pig, NB33 heavy chain, ... | | Authors: | Childers, K.C, Davulcu, O, Haynes, R.M, Lollar, P, Doering, C.B, Coxon, C.H, Spiegel, P.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure of coagulation factor VIII bound to a patient-derived anti-C1 domain antibody inhibitor.
Blood, 142, 2023
|
|
