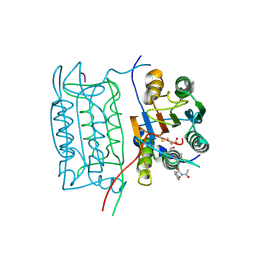
1RWK
 
 | | Crystal structure of human caspase-1 in complex with 3-(2-mercapto-acetylamino)-4-oxo-pentanoic acid | | Descriptor: | 3-(2-MERCAPTO-ACETYLAMINO)-4-OXO-PENTANOIC ACID, Interleukin-1 beta convertase | | Authors: | Romanowski, M.J, Lam, J.W, Fahr, B.T, O'Brien, T. | | Deposit date: | 2003-12-16 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of caspase-1 inhibitors derived from Tethering.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1BMQ
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF INTERLEUKIN-1BETA CONVERTING ENZYME (ICE) WITH A PEPTIDE BASED INHIBITOR, (3S )-N-METHANESULFONYL-3-({1-[N-(2-NAPHTOYL)-L-VALYL]-L-PROLYL }AMINO)-4-OXOBUTANAMIDE | | Descriptor: | (3S)-N-METHANESULFONYL-3-({1-[N-(2-NAPHTOYL)-L-VALYL]-L-PROLYL}AMINO)-4-OXOBUTANAMIDE, PROTEIN (INTERLEUKIN-1 BETA CONVERTASE) | | Authors: | Okamoto, Y, Anan, H, Nakai, E, Morihira, K, Yonetoku, Y, Kurihara, H, Katayama, N, Sakashita, H, Terai, Y, Takeuchi, M, Shibanuma, T, Isomura, Y. | | Deposit date: | 1998-07-24 | | Release date: | 1998-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peptide based interleukin-1 beta converting enzyme (ICE) inhibitors: synthesis, structure activity relationships and crystallographic study of the ICE-inhibitor complex.
Chem.Pharm.Bull., 47, 1999
|
|
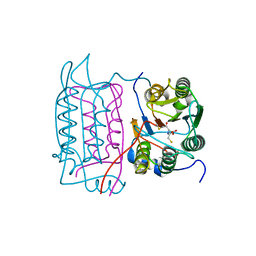
5MTK
 
 | | Crystal structure of human Caspase-1 with (3S,6S,10aS)-N-((2S,3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-6-(isoquinoline-1-carboxamido)-5-oxodecahydropyrrolo[1,2-a]azocine-3-carboxamide (PGE-3935199) | | Descriptor: | (3~{S})-3-[[(3~{S},6~{S},10~{a}~{S})-6-(isoquinolin-1-ylcarbonylamino)-5-oxidanylidene-2,3,6,7,8,9,10,10~{a}-octahydro-1~{H}-pyrrolo[1,2-a]azocin-3-yl]carbonylamino]-4-oxidanyl-butanoic acid, Caspase-1 | | Authors: | Brethon, A, Chantalat, L, Christin, O, Clary, L, Fournier, J.F, Gastreich, M, Harris, C, Pascau, J, Isabet, T, Rodeschin, V, Thoreau, E, Roche, D. | | Deposit date: | 2017-01-09 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Playing against the odds: scaffold hopping from 3D-fragments
To Be Published
|
|
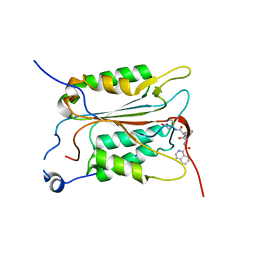
1SC1
 
 | | Crystal structure of an active-site ligand-free form of the human caspase-1 C285A mutant | | Descriptor: | CHLORIDE ION, Interleukin-1 beta convertase | | Authors: | Romanowski, M.J, Scheer, J.M, O'Brien, T, McDowell, R.S. | | Deposit date: | 2004-02-11 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of a ligand-free and malonate-bound human caspase-1: implications for the mechanism of substrate binding.
Structure, 12, 2004
|
|
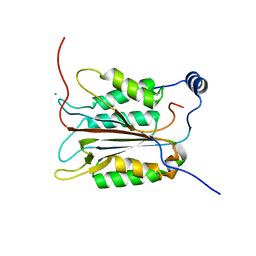
1ICE
 
 | | STRUCTURE AND MECHANISM OF INTERLEUKIN-1BETA CONVERTING ENZYME | | Descriptor: | INTERLEUKIN-1 BETA CONVERTING ENZYME, TETRAPEPTIDE ALDEHYDE | | Authors: | Wilson, K.P, Griffith, J.P, Kim, E.E, Navia, M.A. | | Deposit date: | 1994-09-29 | | Release date: | 1995-07-28 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism of interleukin-1 beta converting enzyme.
Nature, 370, 1994
|
|
3NS7
 
 | |
1RWM
 
 | | Crystal structure of human caspase-1 in complex with 4-oxo-3-[2-(5-{[4-(quinoxalin-2-ylamino)-benzoylamino]-methyl}-thiophen-2-yl)-acetylamino]-pentanoic acid | | Descriptor: | 4-OXO-3-[2-(5-{[4-(QUINOXALIN-2-YLAMINO)-BENZOYLAMINO]-METHYL}-THIOPHEN-2-YL)-ACETYLAMINO]-PENTANOIC ACID, Interleukin-1 beta convertase | | Authors: | Romanowski, M.J, Waal, N.D, Fahr, B.T, O'Brien, T. | | Deposit date: | 2003-12-16 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of caspase-1 inhibitors derived from Tethering.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1IBC
 
 | |
6KN0
 
 | |
1RWW
 
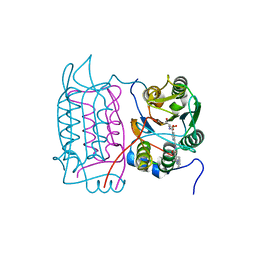 | |
2FQQ
 
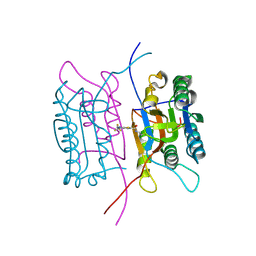 | | Crystal structure of human caspase-1 (Cys285->Ala, Cys362->Ala, Cys364->Ala, Cys397->Ala) in complex with 1-methyl-3-trifluoromethyl-1H-thieno[2,3-c]pyrazole-5-carboxylic acid (2-mercapto-ethyl)-amide | | Descriptor: | 1-METHYL-3-TRIFLUOROMETHYL-1H-THIENO[2,3-C]PYRAZOLE-5-CARBOXYLIC ACID (2-MERCAPTO-ETHYL)-AMIDE, Caspase-1 | | Authors: | Scheer, J.M, Wells, J.A, Romanowski, M.J. | | Deposit date: | 2006-01-18 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A common allosteric site and mechanism in caspases
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
6VIE
 
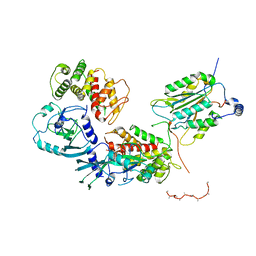 | | Structure of caspase-1 in complex with gasdermin D | | Descriptor: | Caspase-1 subunit p10, Caspase-1 subunit p20, Gasdermin-D | | Authors: | Liu, Z, Xiao, T.S. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Caspase-1 Engages Full-Length Gasdermin D through Two Distinct Interfaces That Mediate Caspase Recruitment and Substrate Cleavage.
Immunity, 53, 2020
|
|
8SV1
 
 | |
7KEU
 
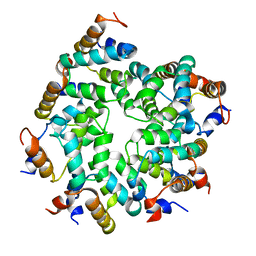 | | Cryo-EM structure of the Caspase-1-CARD:ASC-CARD octamer | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD, Caspase-1 | | Authors: | Hollingsworth, L.R, David, L, Li, Y, Ruan, J, Wu, H. | | Deposit date: | 2020-10-12 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes.
Nat Commun, 12, 2021
|
|
5FNA
 
 | | Cryo-EM reconstruction of caspase-1 CARD | | Descriptor: | Caspase-1 | | Authors: | Li, Y, Lu, A, Schmidt, F.I, Yin, Q, Chen, S, Fu, T.M, Tong, A.B, Ploegh, H.L, Mao, Y, Wu, H. | | Deposit date: | 2015-11-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular Basis of Caspase-1 Polymerization and its Inhibition by a Novel Capping Mechanism
Nat.Struct.Mol.Biol., 23, 2016
|
|
