1Q2B
 
 | | CELLOBIOHYDROLASE CEL7A WITH DISULPHIDE BRIDGE ADDED ACROSS EXO-LOOP BY MUTATIONS D241C AND D249C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, EXOCELLOBIOHYDROLASE I | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2003-07-24 | | Release date: | 2003-11-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D.
J.Mol.Biol., 333, 2003
|
|
1Q2C
 
 | |
1Q2D
 
 | |
1Q2E
 
 | | CELLOBIOHYDROLASE CEL7A WITH LOOP DELETION 245-252 AND BOUND NON-HYDROLYSABLE CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EXOCELLOBIOHYDROLASE I, ... | | Authors: | Stahlberg, J, Harris, M, Jones, T.A. | | Deposit date: | 2003-07-24 | | Release date: | 2003-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A.
A comparison with Phanerochaete chrysosporium Cel7D
J.Mol.Biol., 333, 2003
|
|
1Q2F
 
 | | NMR SOLUTION STRUCTURE OF A PEPTIDE FROM THE MDM-2 BINDING DOMAIN OF THE P53 PROTEIN THAT IS SELECTIVELY CYTOTOXIC TO CANCER CELLS | | Descriptor: | PNC27 | | Authors: | Rosal, R, Pincus, M.R, Brandt-Rauf, P.W, Fine, R.L, Wang, H. | | Deposit date: | 2003-07-24 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a peptide from the mdm-2 binding domain of the p53 protein that is selectively cytotoxic to cancer cells
Biochemistry, 43, 2004
|
|
1Q2H
 
 | | Phenylalanine Zipper Mediates APS Dimerization | | Descriptor: | adaptor protein with pleckstrin homology and src homology 2 domains | | Authors: | Dhe-Paganon, S, Werner, E.D, Nishi, M, Chi, Y.-I, Shoelson, S.E. | | Deposit date: | 2003-07-24 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A phenylalanine zipper mediates APS dimerization.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1Q2I
 
 | | NMR SOLUTION STRUCTURE OF A PEPTIDE FROM THE MDM-2 BINDING DOMAIN OF THE P53 PROTEIN THAT IS SELECTIVELY CYTOTOXIC TO CANCER CELLS | | Descriptor: | PNC27 | | Authors: | Rosal, R, Pincus, M.R, Brandt-Rauf, P.W, Fine, R.L, Wang, H. | | Deposit date: | 2003-07-24 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a peptide from the mdm-2 binding domain of the p53 protein that is selectively cytotoxic to cancer cells
Biochemistry, 43, 2004
|
|
1Q2J
 
 | | Structural basis for tetrodotoxin-resistant sodium channel binding by mu-conotoxin SmIIIA | | Descriptor: | Mu-conotoxin SmIIIA | | Authors: | Keizer, D.W, West, P.J, Lee, E.F, Olivera, B.M, Bulaj, G, Yoshikami, D, Norton, R.S. | | Deposit date: | 2003-07-24 | | Release date: | 2004-02-24 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Structural basis for tetrodotoxin-resistant sodium channel binding by mu-conotoxin SmIIIA.
J.Biol.Chem., 278, 2003
|
|
1Q2K
 
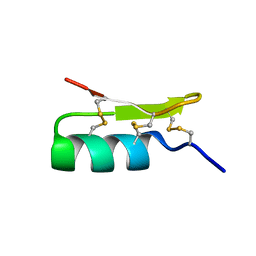 | | Solution structure of BmBKTx1 a new potassium channel blocker from the Chinese Scorpion Buthus martensi Karsch | | Descriptor: | Neurotoxin BmK37 | | Authors: | Cai, Z, Xu, C, Xu, Y, Lu, W, Chi, C.W, Shi, Y, Wu, J. | | Deposit date: | 2003-07-25 | | Release date: | 2003-09-09 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of BmBKTx1, a New BK(Ca)(1) Channel Blocker from the Chinese Scorpion Buthus martensi Karsch(,).
Biochemistry, 43, 2004
|
|
1Q2L
 
 | |
1Q2N
 
 | |
1Q2O
 
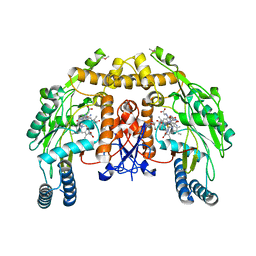 | | Bovine endothelial nitric oxide synthase N368D mutant heme domain dimer with L-N(omega)-nitroarginine-2,4-L-diaminobutyramide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-07-25 | | Release date: | 2004-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1Q2P
 
 | | SHV-1 class A beta-lactamase complexed with penem WAY185229 | | Descriptor: | (6,7-DIHYDRO-5H-CYCLOPENTA[D]IMIDAZO[2,1-B]THIAZOL-2-YL]-4,7-DIHYDRO[1,4]THIAZEPINE-3,6-DICARBOXYLIC ACID, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, beta-lactamase SHV-1 | | Authors: | Nukaga, M, Venkatesan, A.M, Mansour, T.S, Hujer, A, Bonomo, R.A, Knox, J.R. | | Deposit date: | 2003-07-25 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-activity relationship of 6-methylidene penems bearing tricyclic heterocycles as broad-spectrum beta-lactamase inhibitors: crystallographic structures show unexpected binding of 1,4-thiazepine intermediates
J.Med.Chem., 47, 2004
|
|
1Q2Q
 
 | | Enterobacter cloacae GC1 class C beta-lactamase complexed with penem WAY185229 | | Descriptor: | (6,7-DIHYDRO-5H-CYCLOPENTA[D]IMIDAZO[2,1-B]THIAZOL-2-YL]-4,7-DIHYDRO[1,4]THIAZEPINE-3,6-DICARBOXYLIC ACID, GLYCEROL, class C beta-lactamase | | Authors: | Nukaga, M, Venkatesan, A.M, Mansour, T.S, Hujer, A, Bonomo, R.A, Knox, J.R. | | Deposit date: | 2003-07-25 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-activity relationship of 6-methylidene penems bearing tricyclic heterocycles as broad-spectrum beta-lactamase inhibitors: crystallographic structures show unexpected binding of 1,4-thiazepine intermediates
J.Med.Chem., 47, 2004
|
|
1Q2R
 
 | | Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate | | Descriptor: | 9-DEAZAGUANINE, Queuine tRNA-ribosyltransferase, RNA (5'-R(*AP*GP*CP*AP*CP*GP*GP*CP*UP*(N)P*UP*AP*AP*AP*CP*CP*GP*UP*GP*C)-3'), ... | | Authors: | Xie, W, Liu, X, Huang, R.H. | | Deposit date: | 2003-07-25 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate
Nat.Struct.Biol., 10, 2003
|
|
1Q2S
 
 | | Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate | | Descriptor: | 5'-R(*AP*GP*CP*AP*CP*GP*GP*CP*UP*(PQ1)P*UP*AP*AP*AP*CP*CP*GP*UP*GP*C)-3', 9-DEAZAGUANINE, Queuine tRNA-ribosyltransferase, ... | | Authors: | Xie, W, Liu, X, Huang, R.H. | | Deposit date: | 2003-07-25 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate
Nat.Struct.Biol., 10, 2003
|
|
1Q2T
 
 | | Solution structure of d(5mCCTCTCC)4 | | Descriptor: | 5'-D(*(MCY)P*CP*TP*CP*TP*CP*C)-3' | | Authors: | Leroy, J.-L. | | Deposit date: | 2003-07-26 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | T.T pair intercalation and duplex inter-conversion within i-motif tetramers
J.Mol.Biol., 333, 2003
|
|
1Q2U
 
 | | Crystal structure of DJ-1/RS and implication on familial Parkinson's disease | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Huai, Q, Sun, Y, Wang, H, Chin, L.S, Li, L, Robinson, H, Ke, H. | | Deposit date: | 2003-07-26 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of DJ-1/RS and implication on familial Parkinson's disease
Febs Lett., 549, 2003
|
|
1Q2V
 
 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form) | | Descriptor: | SULFATE ION, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Yohda, M, Maruyama, T, Miki, K. | | Deposit date: | 2003-07-26 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
1Q2W
 
 | | X-Ray Crystal Structure of the SARS Coronavirus Main Protease | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3C-like protease | | Authors: | Bonanno, J.B, Fowler, R, Gupta, S, Hendle, J, Lorimer, D, Romero, R, Sauder, J.M, Wei, C.L, Liu, E.T, Burley, S.K, Harris, T. | | Deposit date: | 2003-07-26 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Company Says It Mapped Part of SARS Virus
New York Times, 30 July, 2003
|
|
1Q2X
 
 | | Crystal Structure of the E243D Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae bound with substrate aspartate semialdehyde | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-07-26 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1Q2Y
 
 | | Crystal structure of the protein YJCF from Bacillus subtilis: a member of the GCN5-related N-acetyltransferase superfamily fold | | Descriptor: | similar to hypothetical proteins | | Authors: | Fedorov, A.A, Ramagopal, U.A, Fedorov, E.V, Thirumuruhan, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-07-27 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the protein YJCF from Bacillus subtilis: a member of the GCN5-related N-acetyltransferase superfamily
To be Published
|
|
1Q2Z
 
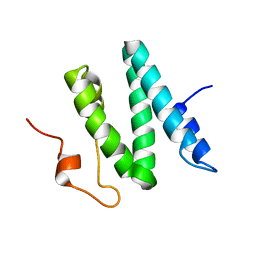 | | The 3D solution structure of the C-terminal region of Ku86 | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Harris, R, Esposito, D, Sankar, A, Maman, J.D, Hinks, J.A, Pearl, L.H, Driscoll, P.C. | | Deposit date: | 2003-07-28 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The 3D Solution Structure of the C-terminal Region of Ku86 (Ku86CTR)
J.Mol.Biol., 335, 2004
|
|
1Q31
 
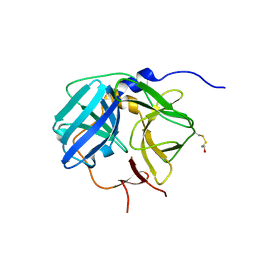 | | Crystal Structure of the Tobacco Etch Virus Protease C151A mutant | | Descriptor: | BETA-MERCAPTOETHANOL, Nuclear inclusion protein A | | Authors: | Nunn, C.M, Djordjevic, S, George, R.R, Urquhart, G.T, Chao, L.H, Tsuchiya, Y. | | Deposit date: | 2003-07-28 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of tobacco etch virus protease shows the protein C terminus bound within the active site.
J.Mol.Biol., 350, 2005
|
|
1Q32
 
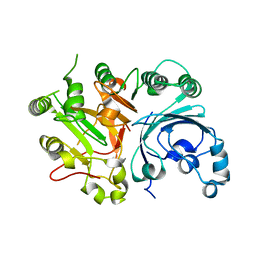 | | Crystal Structure Analysis of the Yeast Tyrosyl-DNA Phosphodiesterase | | Descriptor: | tyrosyl-DNA phosphodiesterase | | Authors: | He, X, Babaoglu, K, Price, A, Nitiss, K.C, Nitiss, J.L, White, S.W. | | Deposit date: | 2003-07-28 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mutation of a conserved active site residue converts tyrosyl-DNA phosphodiesterase I into a DNA topoisomerase I-dependent poison
J.Mol.Biol., 372, 2007
|
|
