1KFE
 
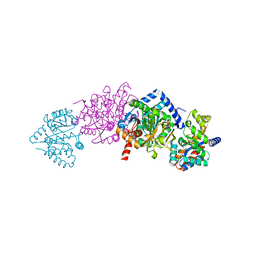 | | CRYSTAL STRUCTURE OF ALPHAT183V MUTANT OF TRYPTOPHAN SYNTHASE FROM SALMONELLA TYPHIMURIUM WITH L-Ser Bound To The Beta Site | | Descriptor: | SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, TRYPTOPHAN SYNTHASE BETA CHAIN, ... | | Authors: | Kulik, V, Weyand, M, Siedel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-11-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | On the Role of AlphaTHR183 in the Allosteric Regulation and Catalytic Mechanism of Tryptophan Synthase
J.Mol.Biol., 324, 2002
|
|
1KFF
 
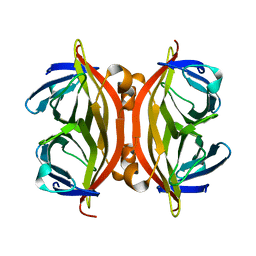 | |
1KFG
 
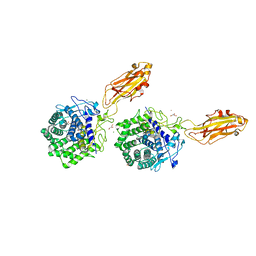 | | The X-ray Crystal Structure of Cel9G from Clostridium cellulolyticum complexed with a Thio-Oligosaccharide Inhibitor | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-thio-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-1-thio-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.-P, Driguez, H, Haser, R. | | Deposit date: | 2001-11-20 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray crystal structure of the multidomain endoglucanase Cel9G from Clostridium cellulolyticum complexed with natural and synthetic cello-olligosaccharides
J.Bacteriol., 185, 2003
|
|
1KFH
 
 | |
1KFI
 
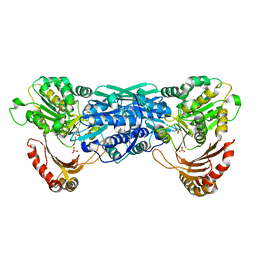 | | Crystal Structure of the Exocytosis-Sensitive Phosphoprotein, pp63/Parafusin (phosphoglucomutase) from Paramecium | | Descriptor: | SULFATE ION, ZINC ION, phosphoglucomutase 1 | | Authors: | Mueller, S, Diederichs, K, Breed, J, Kissmehl, R, Hauser, K, Plattner, H, Welte, W. | | Deposit date: | 2001-11-21 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure analysis of the exocytosis-sensitive phosphoprotein, pp63/parafusin (phosphoglucomutase), from Paramecium reveals significant conformational variability.
J.Mol.Biol., 315, 2002
|
|
1KFJ
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH L-SERINE | | Descriptor: | SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, TRYPTOPHAN SYNTHASE BETA CHAIN, ... | | Authors: | Kulik, V, Weyand, M, Seidel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-11-21 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | On the role of alphaThr183 in the allosteric regulation and catalytic mechanism of tryptophan synthase.
J.Mol.Biol., 324, 2002
|
|
1KFK
 
 | | Crystal structure of Tryptophan Synthase From Salmonella Typhimurium | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Kulik, V, Weyand, M, Seidel, R, Niks, D, Arac, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2001-11-21 | | Release date: | 2003-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | On the role of alphaThr183 in the allosteric regulation and catalytic mechanism of tryptophan synthase.
J.Mol.Biol., 324, 2002
|
|
1KFL
 
 | | Crystal structure of phenylalanine-regulated 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) from E.coli complexed with Mn2+, PEP, and Phe | | Descriptor: | 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase, MANGANESE (II) ION, PHENYLALANINE, ... | | Authors: | Shumilin, I.A, Zhao, C, Bauerle, R, Kretsinger, R.H. | | Deposit date: | 2001-11-21 | | Release date: | 2002-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric inhibition of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase alters the coordination of both substrates.
J.Mol.Biol., 320, 2002
|
|
1KFM
 
 | |
1KFN
 
 | |
1KFO
 
 | | CRYSTAL STRUCTURE OF AN RNA HELIX RECOGNIZED BY A ZINC-FINGER PROTEIN: AN 18 BASE PAIR DUPLEX AT 1.6 RESOLUTION | | Descriptor: | 5'-R(*GP*AP*AP*UP*GP*CP*CP*UP*GP*CP*GP*AP*GP*CP*AP*(5BU)P*CP*CP*C)-3' | | Authors: | Lima, S, Hildenbrand, J, Korostelev, A, Hattman, S, Li, H. | | Deposit date: | 2001-11-21 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of an RNA helix recognized by a zinc-finger protein: an 18-bp duplex at 1.6 A resolution.
RNA, 8, 2002
|
|
1KFP
 
 | | Solution structure of the antimicrobial 18-residue gomesin | | Descriptor: | GOMESIN | | Authors: | Mandard, N, Bulet, P, Caille, A, Daffre, S, Vovelle, F. | | Deposit date: | 2001-11-22 | | Release date: | 2002-04-10 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | The solution structure of gomesin, an antimicrobial cysteine-rich peptide from the spider.
Eur.J.Biochem., 269, 2002
|
|
1KFQ
 
 | | Crystal Structure of Exocytosis-Sensitive Phosphoprotein, pp63/parafusin (Phosphoglucomutse) from Paramecium. OPEN FORM | | Descriptor: | CALCIUM ION, phosphoglucomutase 1 | | Authors: | Mueller, S, Diederichs, K, Breed, J, Kissmehl, R, Hauser, K, Plattner, H, Welte, W. | | Deposit date: | 2001-11-22 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure analysis of the exocytosis-sensitive phosphoprotein, pp63/parafusin (phosphoglucomutase), from Paramecium reveals significant conformational variability.
J.Mol.Biol., 315, 2002
|
|
1KFR
 
 | | Structural plasticity in the eight-helix fold of a trematode hemoglobin | | Descriptor: | Hemoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Milani, M, Pesce, A, Dewilde, S, Ascenzi, P, Moens, L, Bolognesi, M. | | Deposit date: | 2001-11-22 | | Release date: | 2002-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural plasticity in the eight-helix fold of a trematode haemoglobin.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KFS
 
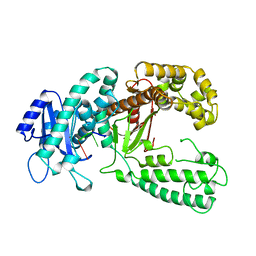 | | DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7) MUTANT/DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*AP*CP*G)-3'), MAGNESIUM ION, PROTEIN (DNA POLYMERASE I KLENOW FRAGMENT (E.C.2.7.7.7)), ... | | Authors: | Brautigam, C.A, Steitz, T.A. | | Deposit date: | 1997-08-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural principles for the inhibition of the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates.
J.Mol.Biol., 277, 1998
|
|
1KFT
 
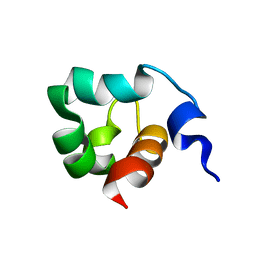 | | Solution Structure of the C-Terminal domain of UvrC from E-coli | | Descriptor: | Excinuclease ABC subunit C | | Authors: | Singh, S, Folkers, G.E, Bonvin, A.M.J.J, Boelens, R, Wechselberger, R, Niztayev, A, Kaptein, R. | | Deposit date: | 2001-11-23 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E.coli
EMBO J., 21, 2002
|
|
1KFU
 
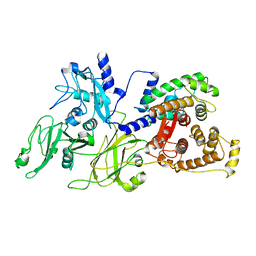 | | Crystal Structure of Human m-Calpain Form II | | Descriptor: | M-CALPAIN LARGE SUBUNIT, M-CALPAIN SMALL SUBUNIT | | Authors: | Strobl, S, Fernandez-Catalan, C, Braun, M, Huber, R, Masumoto, H, Nakagawa, K, Irie, A, Sorimachi, H, Bourenkow, G, Bartunik, H, Suzuki, K, Bode, W. | | Deposit date: | 2001-11-23 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1KFV
 
 | | Crystal Structure of Lactococcus lactis Formamido-pyrimidine DNA Glycosylase (alias Fpg or MutM) Non Covalently Bound to an AP Site Containing DNA. | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*(PDI)P*TP*TP*TP*CP*TP*C)-3', 5'-D(*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*AP*G)-3', Formamido-pyrimidine DNA glycosylase, ... | | Authors: | Serre, L, Pereira de Jesus, K, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2001-11-23 | | Release date: | 2002-06-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the Lactococcus lactis formamidopyrimidine-DNA glycosylase bound to an abasic site analogue-containing DNA.
EMBO J., 21, 2002
|
|
1KFW
 
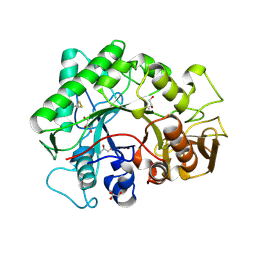 | |
1KFX
 
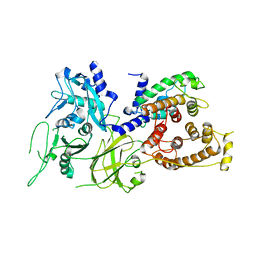 | | Crystal Structure of Human m-Calpain Form I | | Descriptor: | M-CALPAIN LARGE SUBUNIT, M-CALPAIN SMALL SUBUNIT | | Authors: | Strobl, S, Fernandez-Catalan, C, Braun, M, Huber, R, Masumoto, H, Nakagawa, K, Irie, A, Sorimachi, H, Bourenkow, G, Bartunik, H, Suzuki, K, Bode, W. | | Deposit date: | 2001-11-23 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1KFY
 
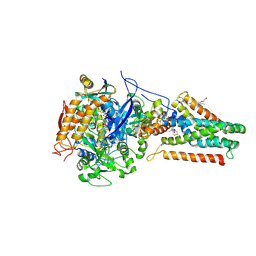 | | QUINOL-FUMARATE REDUCTASE WITH QUINOL INHIBITOR 2-[1-(4-CHLORO-PHENYL)-ETHYL]-4,6-DINITRO-PHENOL | | Descriptor: | 2-[1-(4-CHLORO-PHENYL)-ETHYL]-4,6-DINITRO-PHENOL, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Iverson, T.M, Luna-Chavez, C, Croal, L.R, Cecchini, G, Rees, D.C. | | Deposit date: | 2001-11-24 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the
quinol-binding site.
J.Biol.Chem., 277, 2002
|
|
1KFZ
 
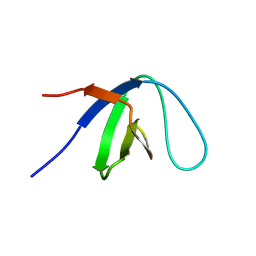 | | Solution Structure of C-terminal Sem-5 SH3 Domain (Ensemble of 16 Structures) | | Descriptor: | SEX MUSCLE ABNORMAL PROTEIN 5 | | Authors: | Ferreon, J.C, Volk, D.E, Luxon, B.A, Gorenstein, D, Hilser, V.J. | | Deposit date: | 2001-11-24 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics and Thermodynamics of the Native State Ensemble
of Sem-5 C-Terminal SH3 Domain
Biochemistry, 42, 2003
|
|
1KG0
 
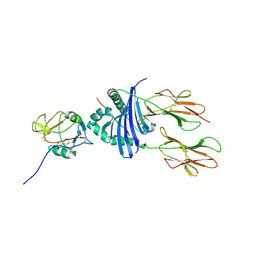 | | Structure of the Epstein-Barr Virus gp42 Protein Bound to the MHC class II Receptor HLA-DR1 | | Descriptor: | Hemagglutinin HA Peptide, MHC class II Receptor HLA-DR1, gp42 Protein | | Authors: | Mullen, M.M, Haan, K.M, Longnecker, R, Jardetzky, T.S. | | Deposit date: | 2001-11-25 | | Release date: | 2002-03-27 | | Last modified: | 2016-11-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1.
Mol.Cell, 9, 2002
|
|
1KG1
 
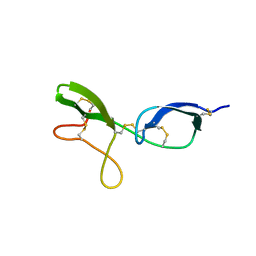 | | NMR structure of the NIP1 elicitor protein from Rhynchosporium secalis | | Descriptor: | Necrosis Inducing Protein 1 | | Authors: | Van't Slot, K.A, Van den Burg, H.A, Kloks, C.P, Hilbers, C.W, Knogge, W, Papavoine, C.H. | | Deposit date: | 2001-11-26 | | Release date: | 2003-11-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Plant Disease Resistance-triggering Protein NIP1 from the Fungus Rhynchosporium secalis Shows a Novel beta-Sheet Fold.
J.Biol.Chem., 278, 2003
|
|
1KG2
 
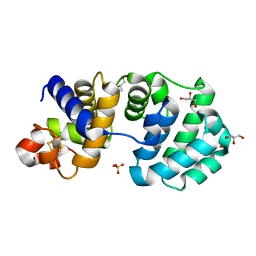 | | Crystal structure of the core fragment of MutY from E.coli at 1.2A resolution | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
