7Z8K
 
 | | Cytoplasmic dynein (A1) bound to BICDR1 | | 分子名称: | ARP1 actin related protein 1 homolog A, BICD family-like cargo adapter 1, Cytoplasmic dynein 1 heavy chain 1, ... | | 著者 | Chaaban, S, Carter, A.P. | | 登録日 | 2022-03-17 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.37 Å) | | 主引用文献 | Structure of dynein-dynactin on microtubules shows tandem adaptor binding.
Nature, 610, 2022
|
|
7Z8F
 
 | |
7Z8J
 
 | | Cytoplasmic dynein (A2) bound to BICDR1 | | 分子名称: | BICD family-like cargo adapter 1, Cytoplasmic dynein 1 heavy chain 1, Cytoplasmic dynein 1 intermediate chain 2, ... | | 著者 | Chaaban, S, Carter, A.P. | | 登録日 | 2022-03-17 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | Structure of dynein-dynactin on microtubules shows tandem adaptor binding.
Nature, 610, 2022
|
|
7Z8I
 
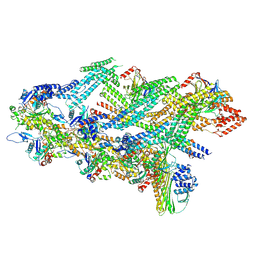 | | The barbed end complex of dynactin bound to BICDR1 and the cytoplasmic dynein tails (A2, B1, B2) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ARP1 actin related protein 1 homolog A, BICD family-like cargo adapter 1, ... | | 著者 | Chaaban, S, Carter, A.P. | | 登録日 | 2022-03-17 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of dynein-dynactin on microtubules shows tandem adaptor binding.
Nature, 610, 2022
|
|
4NTW
 
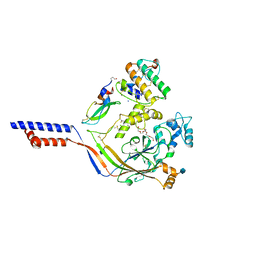 | | Structure of acid-sensing ion channel in complex with snake toxin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid-sensing ion channel 1, Basic phospholipase A2 homolog Tx-beta, ... | | 著者 | Baconguis, I, Bohlen, C.J, Goehring, A, Julius, D, Gouaux, E. | | 登録日 | 2013-12-02 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | X-ray structure of Acid-sensing ion channel 1-snake toxin complex reveals open state of a na(+)-selective channel.
Cell(Cambridge,Mass.), 156, 2014
|
|
4NTY
 
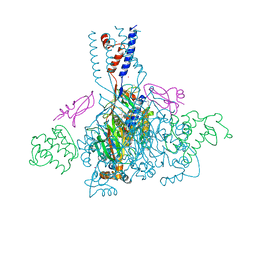 | | Cesium sites in the crystal structure of acid-sensing ion channel in complex with snake toxin | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Acid-sensing ion channel 1, Basic phospholipase A2 homolog Tx-beta, ... | | 著者 | Baconguis, I, Bohlen, C.J, Goehring, A, Julius, D, Gouaux, E. | | 登録日 | 2013-12-02 | | 公開日 | 2014-02-19 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | X-ray structure of Acid-sensing ion channel 1-snake toxin complex reveals open state of a na(+)-selective channel.
Cell(Cambridge,Mass.), 156, 2014
|
|
5VND
 
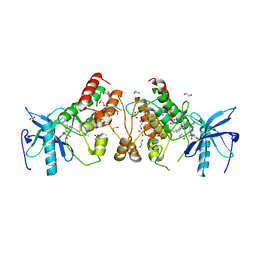 | | Crystal structure of FGFR1-Y563C (FGFR4 surrogate) covalently bound to H3B-6527 | | 分子名称: | 1,2-ETHANEDIOL, Fibroblast growth factor receptor 1, N-{2-[(6-{[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](methyl)amino}pyrimidin-4-yl)amino]-5-(4-ethylpiperazin-1-yl)phenyl}propanamide, ... | | 著者 | Tsai, J.H.C, Reynolds, D, Fekkes, P, Smith, P, Larsen, N.A. | | 登録日 | 2017-04-30 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | H3B-6527 Is a Potent and Selective Inhibitor of FGFR4 in FGF19-Driven Hepatocellular Carcinoma.
Cancer Res., 77, 2017
|
|
5YLZ
 
 | | Cryo-EM Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae at 3.6 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | 登録日 | 2017-10-20 | | 公開日 | 2018-07-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae
Cell, 171, 2017
|
|
6ICZ
 
 | | Cryo-EM structure of a human post-catalytic spliceosome (P complex) at 3.0 angstrom | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | 著者 | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | 登録日 | 2018-09-07 | | 公開日 | 2019-03-13 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6ID0
 
 | | Cryo-EM structure of a human intron lariat spliceosome prior to Prp43 loaded (ILS1 complex) at 2.9 angstrom resolution | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | 登録日 | 2018-09-07 | | 公開日 | 2019-03-13 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
5Y88
 
 | | Cryo-EM structure of the intron-lariat spliceosome ready for disassembly from S.cerevisiae at 3.5 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat, ... | | 著者 | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | 登録日 | 2017-08-20 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structure of an Intron Lariat Spliceosome from Saccharomyces cerevisiae
Cell(Cambridge,Mass.), 171, 2017
|
|
5WSG
 
 | | Cryo-EM structure of the Catalytic Step II spliceosome (C* complex) at 4.0 angstrom resolution | | 分子名称: | 3'-exon-intron, 3'-intron-lariat, 5'-exon, ... | | 著者 | Yan, C, Wan, R, Bai, R, Huang, G, Shi, Y. | | 登録日 | 2016-12-07 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure of a yeast step II catalytically activated spliceosome
Science, 355, 2017
|
|
6J6G
 
 | | Cryo-EM structure of the yeast B*-a2 complex at an average resolution of 3.2 angstrom | | 分子名称: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6ID1
 
 | | Cryo-EM structure of a human intron lariat spliceosome after Prp43 loaded (ILS2 complex) at 2.9 angstrom resolution | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | 登録日 | 2018-09-07 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
5WOV
 
 | |
5WOW
 
 | |
5YZG
 
 | | The Cryo-EM Structure of Human Catalytic Step I Spliceosome (C complex) at 4.1 angstrom resolution | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Zhan, X, Yan, C, Zhang, X, Lei, J, Shi, Y. | | 登録日 | 2017-12-14 | | 公開日 | 2018-08-08 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of a human catalytic step I spliceosome
Science, 359, 2018
|
|
6J6H
 
 | | Cryo-EM structure of the yeast B*-a1 complex at an average resolution of 3.6 angstrom | | 分子名称: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
159D
 
 | | SIDE BY SIDE BINDING OF TWO DISTAMYCIN A DRUGS IN THE MINOR GROOVE OF AN ALTERNATING B-DNA DUPLEX | | 分子名称: | DISTAMYCIN A, DNA (5'-D(*IP*CP*IP*CP*IP*CP*IP*C)-3'), MAGNESIUM ION | | 著者 | Chen, X, Ramakrishnan, B, Rao, S.T, Sundaralingam, M. | | 登録日 | 1994-02-10 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Binding of two distamycin A molecules in the minor groove of an alternating B-DNA duplex.
Nat.Struct.Biol., 1, 1994
|
|
6J6Q
 
 | | Cryo-EM structure of the yeast B*-b2 complex at an average resolution of 3.7 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6J6N
 
 | | Cryo-EM structure of the yeast B*-b1 complex at an average resolution of 3.86 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.86 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
5Z56
 
 | | cryo-EM structure of a human activated spliceosome (mature Bact) at 5.1 angstrom. | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | 登録日 | 2018-01-17 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
1ACY
 
 | |
5XJC
 
 | | Cryo-EM structure of the human spliceosome just prior to exon ligation at 3.6 angstrom | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Zhang, X, Yan, C, Hang, J, Finci, I.L, Lei, J, Shi, Y. | | 登録日 | 2017-04-30 | | 公開日 | 2017-07-05 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | An Atomic Structure of the Human Spliceosome
Cell, 169, 2017
|
|
5Z57
 
 | | Cryo-EM structure of the human activated spliceosome (late Bact) at 6.5 angstrom | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ALANINE, BUD13 homolog, ... | | 著者 | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | 登録日 | 2018-01-17 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
