7DWS
 
 | |
5I5Q
 
 | | Re refinement of 4mwn. | | 分子名称: | DIMETHYL SULFOXIDE, Lysozyme C, NITRATE ION, ... | | 著者 | Helliwell, J.R. | | 登録日 | 2016-02-15 | | 公開日 | 2016-05-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Comment on "Structural dynamics of cisplatin binding to histidine in a protein" [Struct. Dyn. 1, 034701 (2014)].
Struct Dyn, 3, 2016
|
|
5I54
 
 | |
1UU5
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A SOAKED WITH CELLOTETRAOSE | | 分子名称: | ACETATE ION, ENDO-BETA-1,4-GLUCANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Berglund, G.I, Shaw, A, Stahlberg, J, Kenne, L, Driguez, T.H, Mitchinson, C, Sandgren, M. | | 登録日 | 2003-12-15 | | 公開日 | 2004-09-16 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Crystal Complex Structures Reveal How Substrate is Bound in the -4 to the +2 Binding Sites of Humicola Grisea Cel12A
J.Mol.Biol., 342, 2004
|
|
4LS0
 
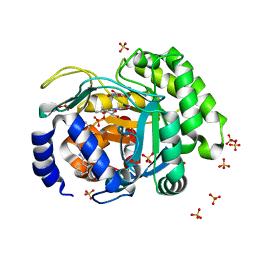 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH01B0033 | | 分子名称: | 2-{(E)-[2-(4-phenyl-1,3-thiazol-2-yl)hydrazinylidene]methyl}benzaldehyde, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | 著者 | Zhu, L, Li, H, Ren, X, Zhu, J. | | 登録日 | 2013-07-21 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DHO1B0033
To be Published
|
|
1HT8
 
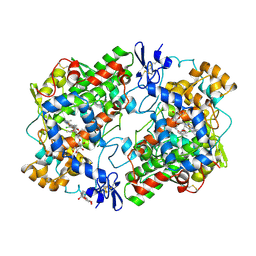 | | THE 2.7 ANGSTROM RESOLUTION MODEL OF OVINE COX-1 COMPLEXED WITH ALCLOFENAC | | 分子名称: | (3-CHLORO-4-PROPOXY-PHENYL)-ACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, PROSTAGLANDIN H2 SYNTHASE-1, ... | | 著者 | Selinsky, B.S, Gupta, K, Sharkey, C.T, Loll, P.J. | | 登録日 | 2000-12-29 | | 公開日 | 2001-04-11 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Structural analysis of NSAID binding by prostaglandin H2 synthase: time-dependent and time-independent inhibitors elicit identical enzyme conformations.
Biochemistry, 40, 2001
|
|
4LT1
 
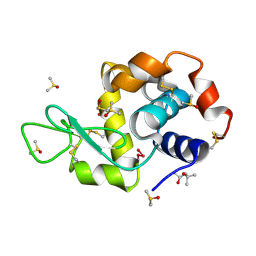 | | HEWL co-crystallised with Carboplatin in non-NaCl conditions: crystal 1 processed using the XDS software package | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, Lysozyme C, ... | | 著者 | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Schreurs, A.M.M, Helliwell, J.R. | | 登録日 | 2013-07-23 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Carboplatin binding to a model protein in non-NaCl conditions to eliminate partial conversion to cisplatin, and the use of different criteria to choose the resolution limit
To be Published
|
|
3MOG
 
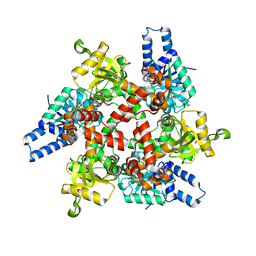 | | Crystal structure of 3-hydroxybutyryl-CoA dehydrogenase from Escherichia coli K12 substr. MG1655 | | 分子名称: | CHLORIDE ION, GLYCEROL, Probable 3-hydroxybutyryl-CoA dehydrogenase | | 著者 | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-04-22 | | 公開日 | 2010-06-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of 3-Hydroxybutyryl-Coa Dehydrogenase from Escherichia Coli K12
To be Published
|
|
3M9S
 
 | | Crystal structure of respiratory complex I from Thermus thermophilus | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | 著者 | Efremov, R.G, Baradaran, R, Sazanov, L.A. | | 登録日 | 2010-03-22 | | 公開日 | 2010-05-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (4.5 Å) | | 主引用文献 | The architecture of respiratory complex I
Nature, 465, 2010
|
|
4Z3M
 
 | | X-ray structure of the adduct formed in the reaction between lysozyme and a platinum(II) Complex with S,O Bidentate Ligands (9b) | | 分子名称: | 1,2-ETHANEDIOL, 3-[2-chloranyl-2-[dimethyl(oxidanyl)-{4}-sulfanyl]-4-ethylsulfanyl-1-oxa-3{3}-thia-2{4}-platinacyclohexa-3,5-dien-6-yl]phenol, DIMETHYL SULFOXIDE, ... | | 著者 | Merlino, A. | | 登録日 | 2015-03-31 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Platinum(II) Complexes with O,S Bidentate Ligands: Biophysical Characterization, Antiproliferative Activity, and Crystallographic Evidence of Protein Binding.
Inorg.Chem., 54, 2015
|
|
1HV0
 
 | | DISSECTING ELECTROSTATIC INTERACTIONS AND THE PH-DEPENDENT ACTIVITY OF A FAMILY 11 GLYCOSIDASE | | 分子名称: | ENDO-1,4-BETA-XYLANASE | | 著者 | Joshi, M.D, Sidhu, G, Nielsen, J.E, Brayer, G.D, Withers, S.G, McIntosh, L.P. | | 登録日 | 2001-01-05 | | 公開日 | 2001-09-14 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Dissecting the electrostatic interactions and pH-dependent activity of a family 11 glycosidase.
Biochemistry, 40, 2001
|
|
4Z40
 
 | | Active site complex BamBC of Benzoyl Coenzyme A reductase as isolated | | 分子名称: | Benzoyl-CoA reductase, putative, IRON/SULFUR CLUSTER, ... | | 著者 | Weinert, T, Kung, J.W, Weidenweber, S, Huwiler, S.G, Boll, M, Ermler, U. | | 登録日 | 2015-04-01 | | 公開日 | 2015-06-24 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural basis of enzymatic benzene ring reduction.
Nat.Chem.Biol., 11, 2015
|
|
1HQX
 
 | | R308K ARGINASE VARIANT | | 分子名称: | ARGINASE, MANGANESE (II) ION | | 著者 | Lavulo, L.T, Sossong Jr, T.M, Brigham-Burke, M.R, Doyle, M.L, Cox, J.D, Christianson, D.W, Ash, D.E. | | 登録日 | 2000-12-20 | | 公開日 | 2001-06-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Subunit-subunit interactions in trimeric arginase. Generation of active monomers by mutation of a single amino acid.
J.Biol.Chem., 276, 2001
|
|
3MDU
 
 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutamate | | 分子名称: | GLYCEROL, N-carbamimidoyl-L-glutamic acid, N-formimino-L-Glutamate Iminohydrolase, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | 登録日 | 2010-03-30 | | 公開日 | 2011-03-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.4003 Å) | | 主引用文献 | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
4ZOA
 
 | | Crystal Structure of beta-glucosidase from Listeria innocua in complex with isofagomine | | 分子名称: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, DI(HYDROXYETHYL)ETHER, Lin1840 protein, ... | | 著者 | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | 登録日 | 2015-05-06 | | 公開日 | 2016-05-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
3MPJ
 
 | | Structure of the glutaryl-coenzyme A dehydrogenase | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, ... | | 著者 | Wischgoll, S, Warkentin, E, Boll, M, Ermler, U. | | 登録日 | 2010-04-27 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for promoting and preventing decarboxylation in glutaryl-coenzyme a dehydrogenases.
Biochemistry, 49, 2010
|
|
4ZPU
 
 | |
1HTY
 
 | | GOLGI ALPHA-MANNOSIDASE II | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | van den Elsen, J.M.H, Kuntz, D.A, Rose, D.R. | | 登録日 | 2001-01-02 | | 公開日 | 2002-01-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure of Golgi alpha-mannosidase II: a target for inhibition of growth and metastasis of cancer cells.
EMBO J., 20, 2001
|
|
3MFW
 
 | |
4Z82
 
 | | Cysteine bound rat cysteine dioxygenase C164S variant at pH 8.1 | | 分子名称: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION | | 著者 | Fellner, M, Tchesnokov, E.P, Siakkou, E, Rutledge, M.T, Kanitz, M, Jameson, G.N.L, Wilbanks, S.M. | | 登録日 | 2015-04-08 | | 公開日 | 2016-06-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Influence of cysteine 164 on active site structure in rat cysteine dioxygenase.
J.Biol.Inorg.Chem., 21, 2016
|
|
4Z85
 
 | |
5I0S
 
 | |
1VBJ
 
 | |
3MIE
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound azide obtained by soaking (50mM NaN3) | | 分子名称: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | 著者 | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | 登録日 | 2010-04-10 | | 公開日 | 2010-11-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.26 Å) | | 主引用文献 | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
4Z9G
 
 | | Crystal structure of human corticotropin-releasing factor receptor 1 (CRF1R) in complex with the antagonist CP-376395 in a hexagonal setting with translational non-crystallographic symmetry | | 分子名称: | 3,6-dimethyl-N-(pentan-3-yl)-2-(2,4,6-trimethylphenoxy)pyridin-4-amine, Corticotropin-releasing factor receptor 1,Lysozyme,Corticotropin-releasing factor receptor 1, OLEIC ACID, ... | | 著者 | Dore, A.S, Bortolato, A, Hollenstein, K, Cheng, R.K.Y, Read, R.J, Marshall, F.H. | | 登録日 | 2015-04-10 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.183 Å) | | 主引用文献 | Decoding Corticotropin-Releasing Factor Receptor Type 1 Crystal Structures.
Curr Mol Pharmacol, 10, 2017
|
|
