5G4O
 
 | | Crystal structure of the p53 cancer mutant Y220C in complex with a trifluorinated derivative of the small molecule stabilizer Phikan083 | | 分子名称: | CELLULAR TUMOR ANTIGEN P53, N,N-dimethyl-1-[9-(2,2,2-trifluoroethyl)-9H-carbazol-3-yl]methanamine, ZINC ION | | 著者 | Joerger, A.C, Bauer, M, Baud, M.G.J, Spencer, J. | | 登録日 | 2016-05-13 | | 公開日 | 2016-06-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Harnessing Fluorine-Sulfur Contacts and Multipolar Interactions for the Design of P53 Mutant Y220C Rescue Drugs.
Acs Chem.Biol., 11, 2016
|
|
8FL9
 
 | |
3RYR
 
 | |
8HPO
 
 | | Cryo-EM structure of a SIN3/HDAC complex from budding yeast | | 分子名称: | Histone deacetylase RPD3, PHOSPHOTHREONINE, POTASSIUM ION, ... | | 著者 | Guo, Z, Zhan, X, Wang, C. | | 登録日 | 2022-12-12 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-07-05 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structure of a SIN3-HDAC complex from budding yeast.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OJ8
 
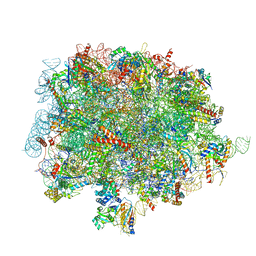 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 1 (native) | | 分子名称: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | 登録日 | 2023-03-24 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8FLC
 
 | |
8FLF
 
 | |
6MWW
 
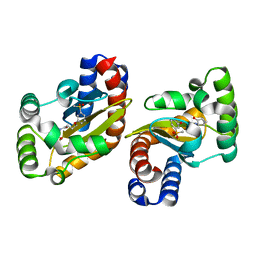 | | LasR LBD:BB0126 complex | | 分子名称: | 4-[3-(methylsulfonyl)phenoxy]-N-[(1R,3R,5R)-2-oxobicyclo[3.1.0]hexan-3-yl]butanamide, Transcriptional regulator LasR | | 著者 | Bassler, B.L, Paczkowski, J.E. | | 登録日 | 2018-10-30 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
1S9K
 
 | |
5YWJ
 
 | | Global regulatory element SarX | | 分子名称: | HTH-type transcriptional regulator SarX | | 著者 | Wang, Q. | | 登録日 | 2017-11-29 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.102 Å) | | 主引用文献 | Crystal structure of SarX from Staphylococcus aureus
To Be Published
|
|
6MWL
 
 | | LasR LBD:mBTL complex | | 分子名称: | 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, Transcriptional regulator LasR | | 著者 | Bassler, B.L, Paczkowski, J.E. | | 登録日 | 2018-10-29 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
6MWZ
 
 | | LasR LBD T75V/Y93F/A127W:BB0126 | | 分子名称: | 4-[3-(methylsulfonyl)phenoxy]-N-[(1S,3S,5S)-2-oxobicyclo[3.1.0]hexan-3-yl]butanamide, ALA-HIS-HIS-HIS-HIS-ALA, Transcriptional regulator LasR | | 著者 | Bassler, B.L, Paczkowski, J.E. | | 登録日 | 2018-10-30 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.657 Å) | | 主引用文献 | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
1MP9
 
 | | TBP from a mesothermophilic archaeon, Sulfolobus acidocaldarius | | 分子名称: | TATA-binding protein | | 著者 | Koike, H, Kawashima-Ohya, Y, Yamasaki, T, Clowney, L, Katsuya, Y, Suzuki, M. | | 登録日 | 2002-09-12 | | 公開日 | 2003-11-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Origins of Protein Stability Revealed by Comparing Crystal Structures of TATA Binding Proteins.
Structure, 12, 2004
|
|
6MWH
 
 | | LasR LBD:BB0020 complex | | 分子名称: | 2-(3-bromophenoxy)-N-[(1S,2S,3R,5S)-2-hydroxybicyclo[3.1.0]hexan-3-yl]acetamide, Transcriptional regulator LasR | | 著者 | Bassler, B.L, Paczkowski, J.E. | | 登録日 | 2018-10-29 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
7LVK
 
 | | Cfr-modified 50S subunit from Escherichia coli | | 分子名称: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Stojkovic, V, Myasnikov, A.G, Frost, A, Fujimori, D.G. | | 登録日 | 2021-02-25 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Investigating antibiotic resistance of a ribosomal-RNA methylating enzyme through directed evolution
To Be Published
|
|
3BR2
 
 | |
3BR0
 
 | |
3BQZ
 
 | |
8J1Z
 
 | |
3BR1
 
 | |
3BTC
 
 | |
3BTJ
 
 | |
3BR6
 
 | |
3BTL
 
 | |
3BR5
 
 | |
