2GPW
 
 | | Crystal Structure of the Biotin Carboxylase Subunit, F363A Mutant, of Acetyl-CoA Carboxylase from Escherichia coli. | | 分子名称: | Biotin carboxylase | | 著者 | Shen, Y, Chou, C.Y, Chang, G.G, Tong, L. | | 登録日 | 2006-04-18 | | 公開日 | 2006-07-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Is dimerization required for the catalytic activity of bacterial biotin carboxylase?
Mol.Cell, 22, 2006
|
|
6DC6
 
 | | Crystal structure of human ubiquitin activating enzyme E1 (Uba1) in complex with ubiquitin | | 分子名称: | MAGNESIUM ION, PYROPHOSPHATE 2-, Ubiquitin, ... | | 著者 | Lv, Z, Yuan, L, Williams, K.M, Atkison, J.H, Olsen, S.K. | | 登録日 | 2018-05-04 | | 公開日 | 2018-10-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | Crystal structure of a human ubiquitin E1-ubiquitin complex reveals conserved functional elements essential for activity.
J. Biol. Chem., 293, 2018
|
|
5LQF
 
 | | CDK1/CyclinB1/CKS2 in complex with NU6102 | | 分子名称: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | 著者 | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B.J, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E, Newell, D.R, Turner, D.M, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Endicott, J.A, Cano, C. | | 登録日 | 2016-08-17 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
7AJU
 
 | | Cryo-EM structure of the 90S-exosome super-complex (state Post-A1-exosome) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | 登録日 | 2020-09-29 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
8T12
 
 | |
8T13
 
 | |
8A3T
 
 | | S. cerevisiae APC/C-Cdh1 complex | | 分子名称: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | 著者 | Barford, D, Vazquez-Fernandez, E, Zhang, Z, Yang, J. | | 登録日 | 2022-06-09 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex
To Be Published
|
|
7ZJX
 
 | | Rabbit 80S ribosome programmed with SECIS and SBP2 | | 分子名称: | 18S rRNA, 28S rRNA, 40S Ribosomal protein eS19, ... | | 著者 | Hilal, T, Simonovic, M, Spahn, C.M.T. | | 登録日 | 2022-04-12 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon.
Science, 376, 2022
|
|
7ZJW
 
 | | Rabbit 80S ribosome as it decodes the Sec-UGA codon | | 分子名称: | 18S rRNA, 28S rRNA, 40S Ribosomal protein eS19, ... | | 著者 | Hilal, T, Simonovic, M, Spahn, C.M.T. | | 登録日 | 2022-04-12 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structure of the mammalian ribosome as it decodes the selenocysteine UGA codon.
Science, 376, 2022
|
|
6MTB
 
 | | Rabbit 80S ribosome with P- and Z-site tRNAs (unrotated state) | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | 著者 | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | 登録日 | 2018-10-19 | | 公開日 | 2018-11-21 | | 最終更新日 | 2019-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
6MTC
 
 | | Rabbit 80S ribosome with Z-site tRNA and IFRD2 (unrotated state) | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | 著者 | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | 登録日 | 2018-10-19 | | 公開日 | 2018-11-21 | | 最終更新日 | 2019-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
7ZW0
 
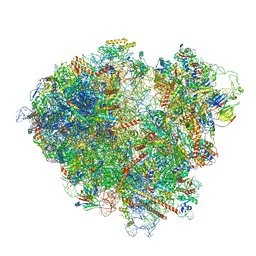 | | FAP-80S Complex - Rotated state | | 分子名称: | 18S ribosomal RNA (RDN18-1), 25S ribosomal RNA (RDN25-1), 40S ribosomal protein S0-A, ... | | 著者 | Ikeuchi, K, Buschauer, R, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2022-05-17 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Sensing of individual stalled 80S ribosomes by Fap1 for nonfunctional rRNA turnover.
Mol.Cell, 82, 2022
|
|
3GZN
 
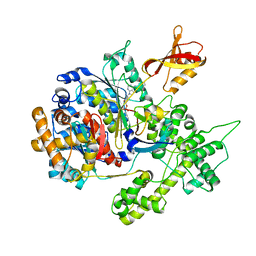 | | Structure of NEDD8-activating enzyme in complex with NEDD8 and MLN4924 | | 分子名称: | NEDD8, NEDD8-activating enzyme E1 catalytic subunit, NEDD8-activating enzyme E1 regulatory subunit, ... | | 著者 | Sintchak, M.D. | | 登録日 | 2009-04-07 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ.
Mol.Cell, 37, 2010
|
|
8RJD
 
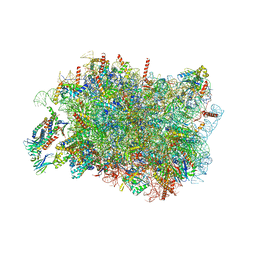 | |
8RJC
 
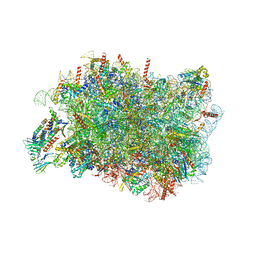 | |
8RJB
 
 | |
3J7O
 
 | | Structure of the mammalian 60S ribosomal subunit | | 分子名称: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | 著者 | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | 登録日 | 2014-08-01 | | 公開日 | 2014-09-03 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
1HON
 
 | | STRUCTURE OF GUANINE NUCLEOTIDE (GPPCP) COMPLEX OF ADENYLOSUCCINATE SYNTHETASE FROM ESCHERICHIA COLI AT PH 6.5 AND 25 DEGREE CELSIUS | | 分子名称: | ADENYLOSUCCINATE SYNTHETASE, AMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Poland, B.W, Hou, Z, Bruns, C, Fromm, H.J, Honzatko, R.B. | | 登録日 | 1996-04-26 | | 公開日 | 1996-11-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Refined crystal structures of guanine nucleotide complexes of adenylosuccinate synthetase from Escherichia coli.
J.Biol.Chem., 271, 1996
|
|
7SYU
 
 | | Structure of the delta dII IRES w/o eIF2 48S initiation complex, closed conformation. Structure 13(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYK
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 5(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYT
 
 | | Structure of the wt IRES w/o eIF2 48S initiation complex, closed conformation. Structure 13(wt) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYH
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 2(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYV
 
 | | Structure of the wt IRES eIF5B-containing pre-48S initiation complex, open conformation. Structure 14(wt) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYO
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head open. Structure 9(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
8P09
 
 | | 48S late-stage initiation complex with non methylated mRNA | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | 著者 | Guca, E, Lima, L.H.F, Boissier, F, Hashem, Y. | | 登録日 | 2023-05-09 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | N 6 -methyladenosine in 5' UTR does not promote translation initiation.
Mol.Cell, 84, 2024
|
|
