7JP1
 
 | |
7K7P
 
 | |
7JZU
 
 | |
7K1O
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-3',5'-Diphosphate | | 分子名称: | 1,2-ETHANEDIOL, 1-(3,5-di-O-phosphono-alpha-L-xylofuranosyl)pyrimidine-2,4(1H,3H)-dione, Uridylate-specific endoribonuclease | | 著者 | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Welk, L, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-09-08 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-3',5'-Diphosphate
To Be Published
|
|
7K9Z
 
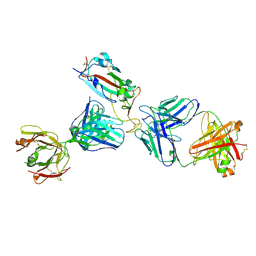 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with the Fab fragments of neutralizing antibodies 298 and 52 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 298 Fab Heavy Chain, 298 Fab Light Chain, ... | | 著者 | Newton, J.C, Kucharska, I, Rujas, E, Cui, H, Julien, J.P. | | 登録日 | 2020-09-29 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Multivalency transforms SARS-CoV-2 antibodies into ultrapotent neutralizers.
Nat Commun, 12, 2021
|
|
7JQC
 
 | | SARS-CoV-2 Nsp1, CrPV IRES and rabbit 40S ribosome complex | | 分子名称: | 40S ribosomal protein S21, 40S ribosomal protein S24, 40S ribosomal protein S26, ... | | 著者 | Yuan, S, Xiong, Y. | | 登録日 | 2020-08-10 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA.
Mol.Cell, 80, 2020
|
|
7JZL
 
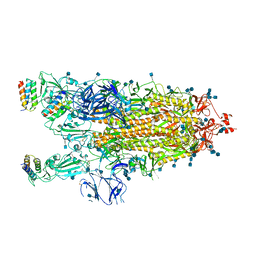 | |
7K43
 
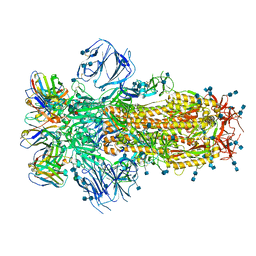 | |
7K0F
 
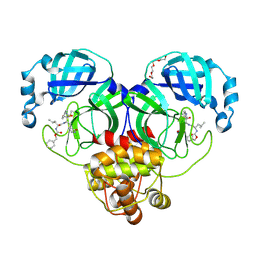 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with a deuterated GC376 alpha-ketoamide analog (compound 5) | | 分子名称: | 3C-like proteinase, N-{(2S,3R)-4-(benzylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-N~2~-[(benzyloxy)carbonyl]-L-leucinamide, TETRAETHYLENE GLYCOL | | 著者 | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Nguyen, H.N, Kim, Y, Chang, K.O, Groutas, W.C. | | 登録日 | 2020-09-04 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JIT
 
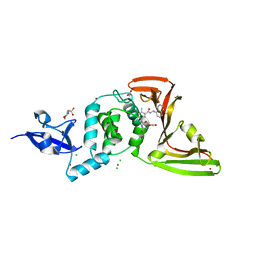 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder495 inhibitor | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(carbamoylcarbamoyl)amino]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-23 | | 公開日 | 2020-08-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JIR
 
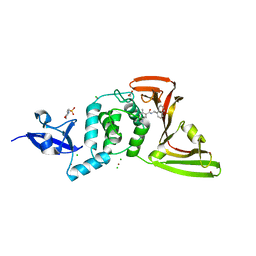 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder457 inhibitor | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, ACETATE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-23 | | 公開日 | 2020-08-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7KDY
 
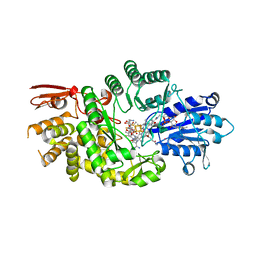 | | Crystal structure of Streptomyces tokunonesis TokK with hydroxycobalamin, 5'-deoxyadenosine, methionine, and (2R)-pantetheinylated carbapenam | | 分子名称: | (2R,3R,5R)-3-{[2-({N-[(2R)-2,4-dihydroxy-3,3-dimethylbutanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-7-oxo-1-azabicyclo[3.2.0]heptane-2-carboxylic acid, 5'-DEOXYADENOSINE, COBALAMIN, ... | | 著者 | Knox, H.L, Booker, S.J, Boal, A.K. | | 登録日 | 2020-10-09 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.939 Å) | | 主引用文献 | Structure of a B 12 -dependent radical SAM enzyme in carbapenem biosynthesis.
Nature, 602, 2022
|
|
7KDX
 
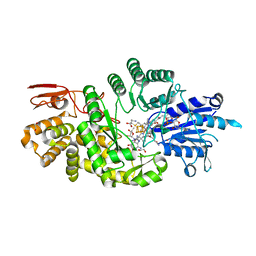 | | Crystal structure of Streptomyces tokunonesis TokK with hydroxycobalamin, 5'-deoxyadenosine, and methionine | | 分子名称: | 1,2-ETHANEDIOL, 5'-DEOXYADENOSINE, CHLORIDE ION, ... | | 著者 | Knox, H.L, Booker, S.J, Boal, A.K. | | 登録日 | 2020-10-09 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.791 Å) | | 主引用文献 | Structure of a B 12 -dependent radical SAM enzyme in carbapenem biosynthesis.
Nature, 602, 2022
|
|
7LB7
 
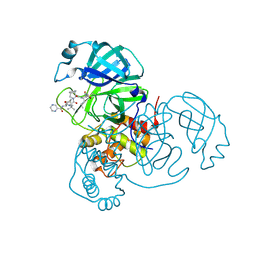 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with Telaprevir | | 分子名称: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A.Y, Kneller, D.W, Coates, L. | | 登録日 | 2021-01-07 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | 主引用文献 | Direct Observation of Protonation State Modulation in SARS-CoV-2 Main Protease upon Inhibitor Binding with Neutron Crystallography.
J.Med.Chem., 64, 2021
|
|
7LG7
 
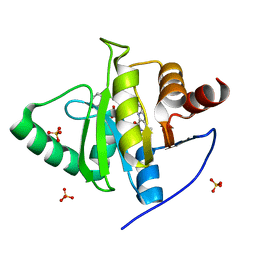 | | Crystal structure of CoV-2 Nsp3 Macrodomain complex with PARG345 | | 分子名称: | 3-[(1,3-dimethyl-2,6-dioxo-2,3,6,9-tetrahydro-1H-purin-8-yl)sulfanyl]-N-{[2-(morpholin-4-yl)ethyl]sulfonyl}propanamide, Non-structural protein 3, SULFATE ION | | 著者 | Arvai, A, Brosey, C.A, Bommagani, S, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | 登録日 | 2021-01-19 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7LLF
 
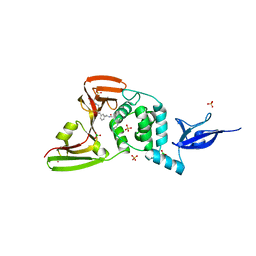 | | SARS-CoV-2 papain-like protease (PLpro) bound to inhibitor XR8-83 | | 分子名称: | 5-[(azetidin-3-yl)amino]-N-[(1R)-1-{3-[5-({[(1R,3S)-3-hydroxycyclopentyl]amino}methyl)thiophen-2-yl]phenyl}ethyl]-2-methylbenzamide, BORIC ACID, GLYCEROL, ... | | 著者 | Ratia, K.M, Xiong, R, Thatcher, G.R. | | 登録日 | 2021-02-03 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures.
Biorxiv, 2021
|
|
7L7K
 
 | |
7LBS
 
 | | SARS-CoV-2 papain-like protease (PLpro) bound to inhibitor XR8-24 | | 分子名称: | 5-[(azetidin-3-yl)amino]-2-methyl-N-[(1R)-1-(3-{5-[(pyrrolidin-1-yl)methyl]thiophen-2-yl}phenyl)ethyl]benzamide, BORIC ACID, GLYCEROL, ... | | 著者 | Ratia, K.M, Xiong, R, Thatcher, G.R. | | 登録日 | 2021-01-08 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures.
Biorxiv, 2021
|
|
7LOS
 
 | | SARS-CoV-2 papain-like protease (PLpro) bound to inhibitor XR8-65 | | 分子名称: | 5-(azetidin-3-ylamino)-2-methyl-~{N}-[(1~{R})-1-[3-[5-[[[(3~{R})-oxolan-3-yl]amino]methyl]thiophen-2-yl]phenyl]ethyl]benzamide, CHLORIDE ION, Non-structural protein 3, ... | | 著者 | Ratia, K.M, Xiong, R, Thatcher, G.R. | | 登録日 | 2021-02-10 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures.
Biorxiv, 2021
|
|
7LLZ
 
 | | SARS-CoV-2 papain-like protease (PLpro) bound to inhibitor XR8-69 | | 分子名称: | N-[(1R)-1-(3-{5-[(acetylamino)methyl]thiophen-2-yl}phenyl)ethyl]-5-[(azetidin-3-yl)amino]-2-methylbenzamide, Non-structural protein 3, SULFATE ION, ... | | 著者 | Ratia, K.M, Xiong, R, Thatcher, G.R. | | 登録日 | 2021-02-04 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures.
Biorxiv, 2021
|
|
7L7F
 
 | |
7LBR
 
 | | SARS-CoV-2 papain-like protease (PLpro) bound to inhibitor XR8-89 | | 分子名称: | 5-[(azetidin-3-yl)amino]-N-[(1R)-1-{3-[5-({[(1S,3R)-3-hydroxycyclopentyl]amino}methyl)thiophen-2-yl]phenyl}ethyl]-2-methylbenzamide, GLYCEROL, Non-structural protein 3, ... | | 著者 | Ratia, K.M, Xiong, R, Thatcher, G.R. | | 登録日 | 2021-01-08 | | 公開日 | 2021-02-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures.
Biorxiv, 2021
|
|
7JOY
 
 | |
7JXD
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | 分子名称: | S2A4 antigen-binding (Fab) fragment | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-27 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JZM
 
 | |
