8CED
 
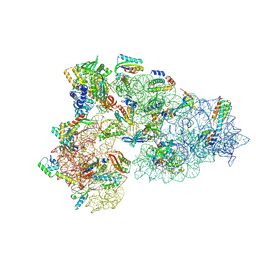 | | Rnase R bound to a 30S degradation intermediate (State I - head-turning) | | 分子名称: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | 著者 | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | 登録日 | 2023-02-01 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.15 Å) | | 主引用文献 | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8CDV
 
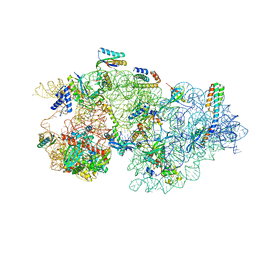 | | Rnase R bound to a 30S degradation intermediate (state II) | | 分子名称: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | 著者 | Paternoga, H, Dimitrova-Paternoga, L, Wilson, D.N. | | 登録日 | 2023-02-01 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.73 Å) | | 主引用文献 | Structural basis of ribosomal 30S subunit degradation by RNase R.
Nature, 626, 2024
|
|
8Q5I
 
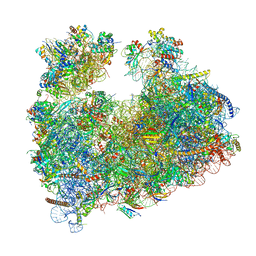 | | Structure of Candida albicans 80S ribosome in complex with cephaeline | | 分子名称: | 18S ribosomal RNA, 25S rRNA, 40S ribosomal protein S0, ... | | 著者 | Kolosova, O, Zgadzay, Y, Stetsenko, A, Atamas, A, Guskov, A, Yusupov, M. | | 登録日 | 2023-08-09 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (2.45 Å) | | 主引用文献 | Structural characterization of cephaeline binding to the eukaryotic ribosome using Cryo-Electron Microscopy
Biopolym Cell, 2023
|
|
1QZD
 
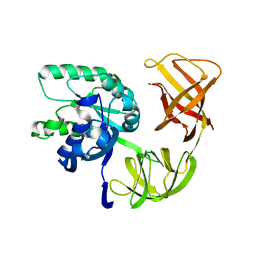 | | EF-Tu.kirromycin coordinates fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | 分子名称: | Elongation factor Tu | | 著者 | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | 登録日 | 2003-09-16 | | 公開日 | 2003-11-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
7SYN
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 8(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYG
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 1(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYH
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 2(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
1QZB
 
 | | Coordinates of the A-site tRNA model fitted into the cryo-EM map of 70S ribosome in the pre-translocational state | | 分子名称: | Phe-tRNA | | 著者 | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | 登録日 | 2003-09-16 | | 公開日 | 2003-11-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1QZA
 
 | | Coordinates of the A/T site tRNA model fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | 分子名称: | Phe-tRNA | | 著者 | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | 登録日 | 2003-09-16 | | 公開日 | 2003-11-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
8P8M
 
 | | Yeast 60S ribosomal subunit, RPL39 deletion | | 分子名称: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | 登録日 | 2023-06-01 | | 公開日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Yeast 60S ribosomal subunit, RPL39 deletion
To Be Published
|
|
6MAT
 
 | | Cryo-EM structure of the essential ribosome assembly AAA-ATPase Rix7 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Rix7 mutant, unknown protein | | 著者 | Lo, Y.H, Sobhany, M, Hsu, A.L, Ford, B.L, Krahn, J.M, Borgnia, M.J, Stanley, R.E. | | 登録日 | 2018-08-28 | | 公開日 | 2019-02-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Cryo-EM structure of the essential ribosome assembly AAA-ATPase Rix7.
Nat Commun, 10, 2019
|
|
8P4V
 
 | | 80S yeast ribosome in complex with HaterumaimideQ | | 分子名称: | (3~{R})-3-[(1~{S})-2-[(1~{R},3~{S},4~{a}~{S},8~{a}~{S})-5,5,8~{a}-trimethyl-2-methylidene-3-oxidanyl-3,4,4~{a},6,7,8-hexahydro-1~{H}-naphthalen-1-yl]-1-oxidanyl-ethyl]pyrrolidine-2,5-dione, 18S ribosomal RNA, 25S ribosomal RNA, ... | | 著者 | Terrosu, S, Yusupov, M. | | 登録日 | 2023-05-23 | | 公開日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.16 Å) | | 主引用文献 | Role of Protein eL42 in the Ribosome Binding of Synthetic Analogues of Natural Lissoclimides
To Be Published
|
|
7OZS
 
 | | Structure of the hexameric 5S RNP from C. thermophilum | | 分子名称: | 5S rRNA, 60S ribosomal protein l5-like protein, Putative ribosomal protein, ... | | 著者 | Castillo, N, Thoms, M, Flemming, D, Hammaren, H.M, Buschauer, R, Ameismeier, M, Bassler, J, Beck, M, Beckmann, R, Hurt, E. | | 登録日 | 2021-06-28 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways.
Nat.Struct.Mol.Biol., 2023
|
|
6YS3
 
 | | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide | | 分子名称: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Schulte, L, Reitz, J, Kudlinzki, D, Hodirnau, V.V, Frangakis, A, Schwalbe, H. | | 登録日 | 2020-04-20 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.58 Å) | | 主引用文献 | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide
Nat Commun, 2020
|
|
4ZC7
 
 | | Paromomycin bound to a leishmanial ribosomal A-site | | 分子名称: | PAROMOMYCIN, RNA duplex | | 著者 | Shalev, M, Rozenberg, H, Jaffe, C.L, Adir, N, Baasov, T. | | 登録日 | 2015-04-15 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.041 Å) | | 主引用文献 | Structural basis for selective targeting of leishmanial ribosomes: aminoglycoside derivatives as promising therapeutics.
Nucleic Acids Res., 43, 2015
|
|
5BY8
 
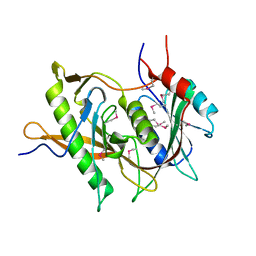 | | The structure of Rpf2-Rrs1 explains its role in ribosome biogenesis | | 分子名称: | Rpf2, Rrs1 | | 著者 | Kharde, S, Calvino, F.R, Gumiero, A, Wild, K, Sinning, I. | | 登録日 | 2015-06-10 | | 公開日 | 2015-07-08 | | 最終更新日 | 2015-08-26 | | 実験手法 | X-RAY DIFFRACTION (1.515 Å) | | 主引用文献 | The structure of Rpf2-Rrs1 explains its role in ribosome biogenesis.
Nucleic Acids Res., 43, 2015
|
|
1C2X
 
 | |
1RY1
 
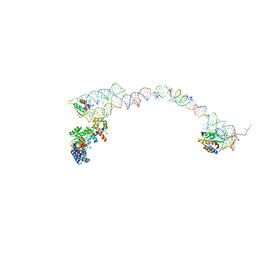 | | Structure of the signal recognition particle interacting with the elongation-arrested ribosome | | 分子名称: | SRP Alu domain, SRP RNA, SRP S domain, ... | | 著者 | Halic, M, Becker, T, Pool, M.R, Spahn, C.M, Grassucci, R.A, Frank, J, Beckmann, R. | | 登録日 | 2003-12-19 | | 公開日 | 2004-04-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (12 Å) | | 主引用文献 | Structure of the signal recognition particle interacting with the elongation-arrested ribosome
Nature, 427, 2004
|
|
2MEZ
 
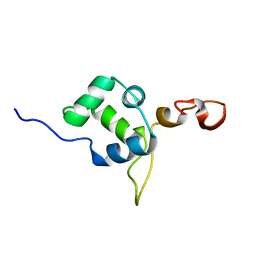 | | Flexible anchoring of archaeal MBF1 on ribosomes suggests role as recruitment factor | | 分子名称: | Multiprotein Bridging Factor (MBP-like) | | 著者 | Launay, H, Blombarch, F, Camilloni, C, Vendruscolo, M, van des Oost, J, Christodoulou, J. | | 登録日 | 2013-10-03 | | 公開日 | 2014-06-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Archaeal MBF1 binds to 30S and 70S ribosomes via its helix-turn-helix domain.
Biochem.J., 462, 2014
|
|
1RQV
 
 | | Spatial model of L7 dimer from E.coli with one hinge region in helical state | | 分子名称: | 50S ribosomal protein L7/L12 | | 著者 | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | 登録日 | 2003-12-07 | | 公開日 | 2004-03-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | From structure and dynamics of protein L7/L12 to molecular switching in ribosome
J.Biol.Chem., 279, 2004
|
|
4PDB
 
 | |
2XDV
 
 | | Crystal Structure of the Catalytic Domain of FLJ14393 | | 分子名称: | 1,2-ETHANEDIOL, CADMIUM ION, MANGANESE (II) ION, ... | | 著者 | Krojer, T, Muniz, J.R.C, Ng, S.S, Pilka, E, Guo, K, Pike, A.C.W, Filippakopoulos, P, Knapp, S, Kavanagh, K.L, Gileadi, O, Bunkoczi, G, Yue, W.W, Niesen, F, Sobott, F, Fedorov, O, Savitsky, P, Kochan, G, Daniel, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U. | | 登録日 | 2010-05-07 | | 公開日 | 2010-05-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
1MOM
 
 | | CRYSTAL STRUCTURE OF MOMORDIN, A TYPE I RIBOSOME INACTIVATING PROTEIN FROM THE SEEDS OF MOMORDICA CHARANTIA | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MOMORDIN | | 著者 | Husain, J, Tickle, I.J, Wood, S.P. | | 登録日 | 1994-03-04 | | 公開日 | 1994-05-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Crystal structure of momordin, a type I ribosome inactivating protein from the seeds of Momordica charantia.
FEBS Lett., 342, 1994
|
|
3IZ4
 
 | |
3MOJ
 
 | |
