5JER
 
 | | Structure of Rotavirus NSP1 bound to IRF-3 | | 分子名称: | Interferon regulatory factor 3, Rotavirus NSP1 peptide | | 著者 | Zhao, B, Li, P. | | 登録日 | 2016-04-18 | | 公開日 | 2016-06-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.913 Å) | | 主引用文献 | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JEJ
 
 | | Phosphorylated STING in complex with IRF-3 CTD | | 分子名称: | Interferon regulatory factor 3, Stimulator of interferon genes protein | | 著者 | Li, P, Shu, C. | | 登録日 | 2016-04-18 | | 公開日 | 2016-06-15 | | 最終更新日 | 2016-06-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for concerted recruitment and activation of IRF-3 by innate immune adaptor proteins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3B0D
 
 | | Crystal structure of the chicken CENP-T histone fold/CENP-W complex, crystal form II | | 分子名称: | CITRIC ACID, Centromere protein T, Centromere protein W | | 著者 | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | 登録日 | 2011-06-08 | | 公開日 | 2012-03-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.197 Å) | | 主引用文献 | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold.
Cell(Cambridge,Mass.), 148, 2012
|
|
1ORC
 
 | |
5H99
 
 | |
5H9I
 
 | | Crystal structure of Geobacter metallireducens SMUG1 with xanthine | | 分子名称: | BETA-MERCAPTOETHANOL, GLYCEROL, Geobacter metallireducens SMUG1, ... | | 著者 | Xie, W, Cao, W, Zhang, Z, Shen, J. | | 登録日 | 2015-12-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.501 Å) | | 主引用文献 | Structural Basis of Substrate Specificity in Geobacter metallireducens SMUG1
Acs Chem.Biol., 11, 2016
|
|
1NQ2
 
 | | Two RTH Mutants with Impaired Hormone Binding | | 分子名称: | ARSENIC, SODIUM ION, Thyroid hormone receptor beta-1, ... | | 著者 | Huber, B.R, Sandler, B, West, B.L, Cunha-Lima, S.T, Nguyen, H.T, Apriletti, J.W, Baxter, J.D, Fletterick, R.J. | | 登録日 | 2003-01-21 | | 公開日 | 2003-04-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Two resistance to thyroid hormone mutants with impaired hormone binding
Mol.Endocrinol., 17, 2003
|
|
3C28
 
 | |
8B3I
 
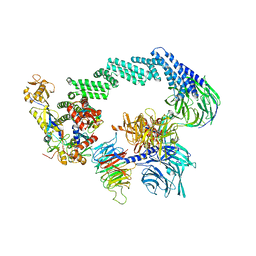 | | CRL4CSA-E2-Ub (state 2) | | 分子名称: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | 著者 | Kokic, G, Cramer, P. | | 登録日 | 2022-09-16 | | 公開日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | C(N)RL4CSA-E2-Ub (state 2)
To Be Published
|
|
6LW3
 
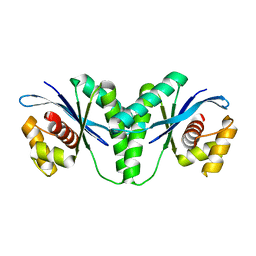 | | Crystal structure of RuvC from Pseudomonas aeruginosa | | 分子名称: | Crossover junction endodeoxyribonuclease RuvC | | 著者 | Hu, Y, He, Y, Lin, Z. | | 登録日 | 2020-02-07 | | 公開日 | 2020-02-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Biochemical and structural characterization of the Holliday junction resolvase RuvC from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
5F4N
 
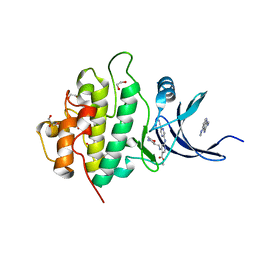 | | Multi-parameter lead optimization to give an oral CHK1 inhibitor clinical candidate: (R)-5-((4-((morpholin-2-ylmethyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)pyrazine-2-carbonitrile (CCT245737) | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Serine/threonine-protein kinase Chk1, ... | | 著者 | Collins, I, Garrett, M.D, van Montfort, R, Osborne, J.D, Matthews, T.P, McHardy, T, Proisy, N, Cheung, K.J, Lainchbury, M, Brown, N, Walton, M.I, Eve, P.D, Boxall, K.J, Hayes, A, Henley, A.T, Valenti, M.R, De Haven Brandon, A.K, Box, G, Westwood, I.M, Jamin, Y, Robinson, S.P, Leonard, P, Reader, J.C, Aherne, G.W, Raynaud, F.I, Eccles, S.A. | | 登録日 | 2015-12-03 | | 公開日 | 2016-05-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Multiparameter Lead Optimization to Give an Oral Checkpoint Kinase 1 (CHK1) Inhibitor Clinical Candidate: (R)-5-((4-((Morpholin-2-ylmethyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)pyrazine-2-carbonitrile (CCT245737).
J.Med.Chem., 59, 2016
|
|
6B1G
 
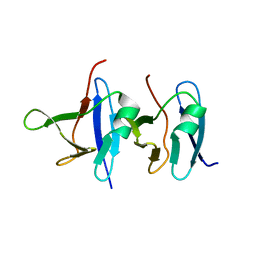 | | Solution structure of TDP-43 N-terminal domain dimer. | | 分子名称: | TAR DNA-binding protein 43, S48E Mutant, Y4R Mutant | | 著者 | Naik, M.T, Wang, A, Conicella, A, Fawzi, N.L. | | 登録日 | 2017-09-18 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing.
EMBO J., 37, 2018
|
|
6M3G
 
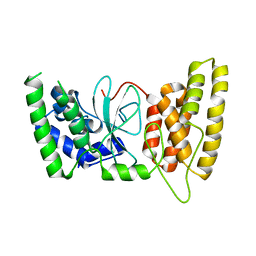 | | Crystal structure of human HPF1 | | 分子名称: | Histone PARylation factor 1 | | 著者 | Sun, F.H, Yun, C.H. | | 登録日 | 2020-03-03 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones.
Nat Commun, 12, 2021
|
|
6M3H
 
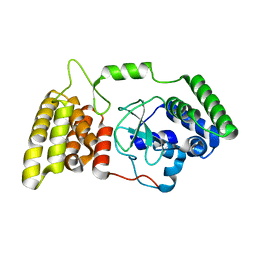 | | Crystal structure of mouse HPF1 | | 分子名称: | Histone PARylation factor 1 | | 著者 | Sun, F.H, Yun, C.H. | | 登録日 | 2020-03-03 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones.
Nat Commun, 12, 2021
|
|
4Y13
 
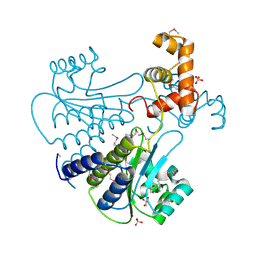 | | SdiA in complex with octanoyl-rac-glycerol | | 分子名称: | (2S)-2,3-dihydroxypropyl octanoate, GLYCEROL, SULFATE ION, ... | | 著者 | Nguyen, N.X, Nguyen, Y, Sperandio, V, Jiang, Y. | | 登録日 | 2015-02-06 | | 公開日 | 2015-04-08 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (3.096 Å) | | 主引用文献 | Structural and Mechanistic Roles of Novel Chemical Ligands on the SdiA Quorum-Sensing Transcription Regulator.
Mbio, 6, 2015
|
|
1G18
 
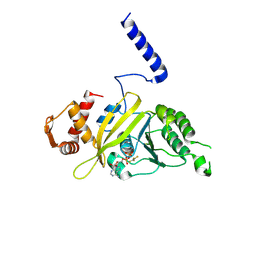 | | RECA-ADP-ALF4 COMPLEX | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, RECA PROTEIN, TETRAFLUOROALUMINATE ION | | 著者 | Datta, S, Prabu, M.M, Vaze, M.B, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2000-10-11 | | 公開日 | 2000-12-11 | | 最終更新日 | 2018-04-04 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Crystal structures of Mycobacterium tuberculosis RecA and its complex with ADP-AlF(4): implications for decreased ATPase activity and molecular aggregation
Nucleic Acids Res., 28, 2000
|
|
5Y81
 
 | | NuA4 TEEAA sub-complex | | 分子名称: | Actin, Actin-related protein 4, Chromatin modification-related protein EAF1, ... | | 著者 | Wang, X, Cai, G. | | 登録日 | 2017-08-18 | | 公開日 | 2018-04-18 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Architecture of the Saccharomyces cerevisiae NuA4/TIP60 complex
Nat Commun, 9, 2018
|
|
8Q1X
 
 | | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | 著者 | Roske, Y, Daumke, O, Damme, M. | | 登録日 | 2023-08-01 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation.
Nucleic Acids Res., 52, 2024
|
|
5C8D
 
 | |
5Y7Y
 
 | | Crystal structure of AhRR/ARNT complex | | 分子名称: | Aryl hydrocarbon receptor nuclear translocator, Aryl hydrocarbon receptor repressor, GLYCEROL | | 著者 | Sakurai, S, Shimizu, T, Ohto, U. | | 登録日 | 2017-08-18 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of the AhRR-ARNT heterodimer reveals the structural basis of the repression of AhR-mediated transcription.
J. Biol. Chem., 292, 2017
|
|
4P78
 
 | | HicA3 and HicB3 toxin-antitoxin complex | | 分子名称: | GLYCEROL, HicA3 Toxin, HicB3 antitoxin | | 著者 | Li de la Sierra-Gallay, I, Bibi-Triki, S, van Tilbeurgh, H, Lazar, N, Pradel, E. | | 登録日 | 2014-03-26 | | 公開日 | 2014-08-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Functional and Structural Analysis of HicA3-HicB3, a Novel Toxin-Antitoxin System of Yersinia pestis.
J.Bacteriol., 196, 2014
|
|
4P7D
 
 | | Antitoxin HicB3 crystal structure | | 分子名称: | Antitoxin HicB3, CHLORIDE ION | | 著者 | Li de la Sierra-Gallay, I, Bibi-Triki, S, van Tilbeurgh, H, Lazar, N, Pradel, E. | | 登録日 | 2014-03-27 | | 公開日 | 2014-08-27 | | 最終更新日 | 2015-02-04 | | 実験手法 | X-RAY DIFFRACTION (2.781 Å) | | 主引用文献 | Functional and Structural Analysis of HicA3-HicB3, a Novel Toxin-Antitoxin System of Yersinia pestis.
J.Bacteriol., 196, 2014
|
|
7K7T
 
 | | Crystal structure of human MORC4 ATPase-CW in complex with AMPPNP | | 分子名称: | Isoform 3 of MORC family CW-type zinc finger protein 4, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Klein, B.J, Tencer, A.H, Kutateladze, T.G. | | 登録日 | 2020-09-24 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Molecular mechanism of the MORC4 ATPase activation.
Nat Commun, 11, 2020
|
|
8Q1K
 
 | | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | 著者 | Roske, Y, Daumke, O, Damme, M. | | 登録日 | 2023-07-31 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation.
Nucleic Acids Res., 52, 2024
|
|
1G19
 
 | | STRUCTURE OF RECA PROTEIN | | 分子名称: | PHOSPHATE ION, RECA PROTEIN | | 著者 | Datta, S, Prabu, M.M, Vaze, M.B, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2000-10-11 | | 公開日 | 2000-12-11 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structures of Mycobacterium tuberculosis RecA and its complex with ADP-AlF(4): implications for decreased ATPase activity and molecular aggregation
Nucleic Acids Res., 28, 2000
|
|
