6WHL
 
 | |
9CA1
 
 | | Human TOP3B-TDRD3 core complex in DNA religation state | | 分子名称: | DNA (5'-D(*AP*TP*T)-3'), DNA (5'-D(P*TP*AP*CP*TP*AP*AP*A)-3'), DNA topoisomerase 3-beta-1, ... | | 著者 | Yang, X, Chen, X, Yang, W, Pommier, Y. | | 登録日 | 2024-06-16 | | 公開日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Structural Insights into human Topoisomerase 3-beta DNA and RNA catalytic cycles and topo-gate dynamics
To Be Published
|
|
8XON
 
 | |
8YE5
 
 | |
8W33
 
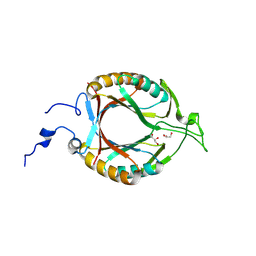 | | Structure of McrD (methyl-coenzyme M reductase operon protein D) from Methanomassiliicoccus luminyensis | | 分子名称: | GLYCEROL, McrD (methyl-coenzyme M reductase operon protein D) | | 著者 | Sutherland-Smith, A.J, Carbone, V, Schofield, L.R, Ronimus, R.S. | | 登録日 | 2024-02-21 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-08-21 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | The crystal structure of methanogen McrD, a methyl-coenzyme M reductase-associated protein.
Febs Open Bio, 14, 2024
|
|
8XUD
 
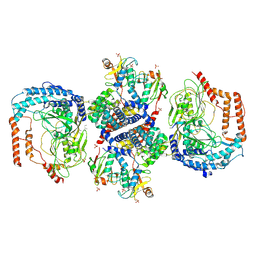 | |
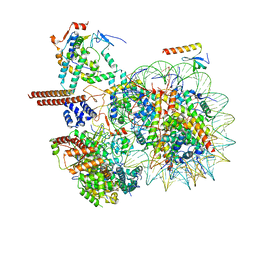
8W9F
 
 | |
6XTG
 
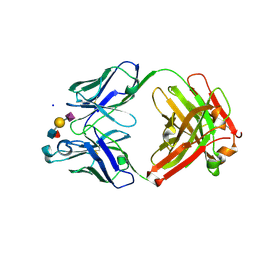 | | Ab 1116NS19.9 bound to CA19-9 | | 分子名称: | Heavy chain, Light chain, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Diskin, R, Borenstein-Katz, A. | | 登録日 | 2020-01-16 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
8VY6
 
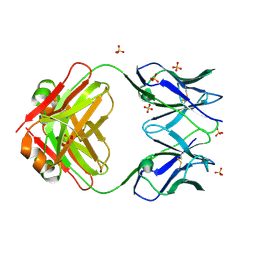 | | Murine light chain dimer | | 分子名称: | 6A8 light chain, SULFATE ION | | 著者 | Kapingidza, A.B, Dolamore, C, Hyduke, N.P, Easly, W, Chivv, C, Pomes, A, Chruszcz, M. | | 登録日 | 2024-02-07 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural, Biophysical, and Computational Studies of a Murine Light Chain Dimer.
Molecules, 29, 2024
|
|
8XLD
 
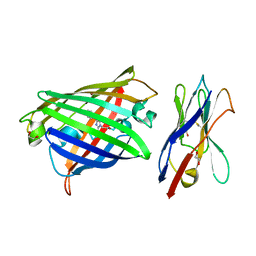 | | Structure of the GFP:GFP-nanobody complex from Biortus. | | 分子名称: | 1,2-ETHANEDIOL, Nanobody(Staygold-S2G10)-Nanobody(Staygold-S4F1), ZINC ION, ... | | 著者 | Wang, F, Cheng, W, Yuan, Z, Lin, D, Bao, C. | | 登録日 | 2023-12-25 | | 公開日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the GFP:GFP-nanobody complex from Biortus.
To Be Published
|
|
8W9E
 
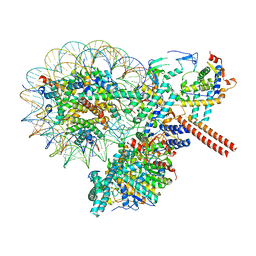 | |
8ZMO
 
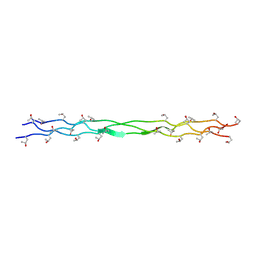 | | Structure of a triple-helix region of human Collagen type XVII from Trautec | | 分子名称: | collagen type XVII | | 著者 | Chu, Y, Zhai, Y, Fan, X, Fu, S, Li, J, Wu, X, Cai, H, Wang, X, Li, D, Feng, P, Cao, K, Qian, S. | | 登録日 | 2024-05-23 | | 公開日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure of a triple-helix region of human Collagen type XVII from Trautec
To Be Published
|
|
8YKW
 
 | | Cryo-EM structure of succinate receptor SUCR1 bound to succinic acid | | 分子名称: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | 登録日 | 2024-03-05 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Molecular basis of ligand recognition and activation of the human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
8X40
 
 | |
8WM4
 
 | |
8Z90
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription initiation conformation 2 | | 分子名称: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | 著者 | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | 登録日 | 2024-04-22 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8XBY
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the blunt end of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
9C4O
 
 | |
8XQU
 
 | | The Crystal Structure of ClpC1-NTD from Biortus. | | 分子名称: | 1,2-ETHANEDIOL, ATP-dependent Clp protease ATP-binding subunit ClpC1, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wang, F, Cheng, W, Lv, Z, Ju, C, Ni, C. | | 登録日 | 2024-01-05 | | 公開日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Crystal Structure of ClpC1-NTD from Biortus.
To Be Published
|
|
8XAW
 
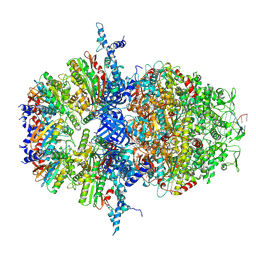 | |
8YZK
 
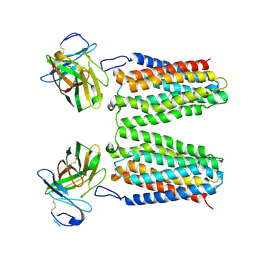 | |
8W1L
 
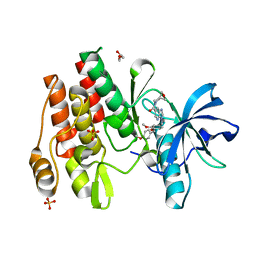 | |
8YRP
 
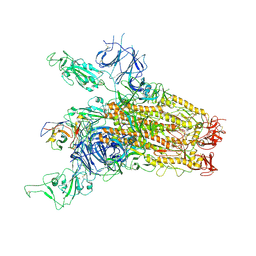 | | SARS-CoV-2 Delta Spike in complex with JM-1A | | 分子名称: | JM-1A Heavy Chain, JM-1A Light Chain, Spike glycoprotein | | 著者 | Nguyen, V.H.T, Chen, X. | | 登録日 | 2024-03-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8WZ2
 
 | | Structure of 26RFa-pyroglutamylated RFamide peptide receptor complex | | 分子名称: | G-alpha q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Jin, S, Li, X, Xu, Y, Guo, S, Wu, C, Zhang, H, Yuan, Q, Xu, H.E, Xie, X, Jiang, Y. | | 登録日 | 2023-11-01 | | 公開日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Structural basis for recognition of 26RFa by the pyroglutamylated RFamide peptide receptor.
Cell Discov, 10, 2024
|
|
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
