7EQV
 
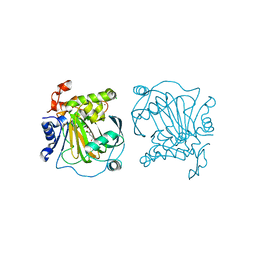 | | Crystal structure of JMJD2A complexed with 3,4-dihydroxybenzoic acid | | 分子名称: | 3,4-DIHYDROXYBENZOIC ACID, CHLORIDE ION, Lysine-specific demethylase 4A, ... | | 著者 | Fang, W.-K, Yang, S.-M, Wang, W.-C. | | 登録日 | 2021-05-04 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Natural product myricetin is a pan-KDM4 inhibitor which with poly lactic-co-glycolic acid formulation effectively targets castration-resistant prostate cancer.
J.Biomed.Sci., 29, 2022
|
|
7EQH
 
 | |
3WLJ
 
 | |
3ZL5
 
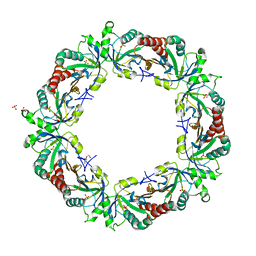 | | Crystal structure of Schistosoma mansoni Peroxiredoxin I C48S mutant with one decamer in the ASU | | 分子名称: | DI(HYDROXYETHYL)ETHER, PEROXIREDOXIN I, SULFATE ION | | 著者 | Saccoccia, F, Angelucci, F, Ardini, M, Boumis, G, Brunori, M, DiLeandro, L, Ippoliti, R, Miele, A.E, Natoli, G, Scotti, S, Bellelli, A. | | 登録日 | 2013-01-28 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.493 Å) | | 主引用文献 | Switching between the Alternative Structures and Functions of a 2-Cys Peroxiredoxin, by Site-Directed Mutagenesis
J.Mol.Biol., 425, 2013
|
|
3ZNJ
 
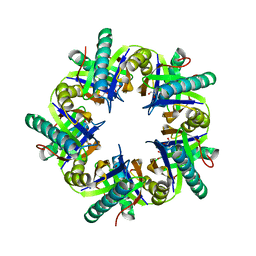 | | Crystal structure of unliganded ClcF from R.opacus 1CP in crystal form 1. | | 分子名称: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION | | 著者 | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | 登録日 | 2013-02-14 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
8FDT
 
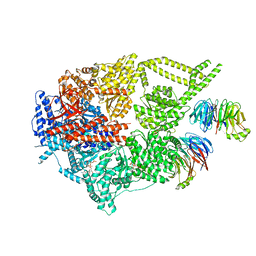 | | Engineered human dynein motor domain in the microtubule-unbound state with LIS1 complex in the buffer containing ATP-Vi | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1,Serine--tRNA ligase, ... | | 著者 | Ton, W, Wang, Y, Chai, P. | | 登録日 | 2022-12-04 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Microtubule-binding-induced allostery triggers LIS1 dissociation from dynein prior to cargo transport.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6FG3
 
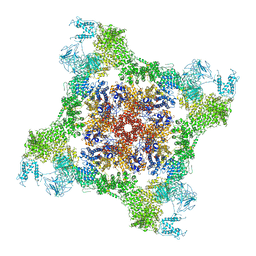 | |
6FKG
 
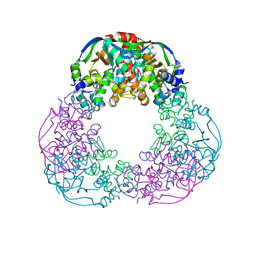 | | Crystal structure of the M.tuberculosis MbcT-MbcA toxin-antitoxin complex. | | 分子名称: | GLYCEROL, Rv1989c (MbcT), Rv1990c (MbcA) | | 著者 | Freire, D.M, Cianci, M, Pogenberg, V, Schneider, T.R, Wilmanns, M, Parret, A.H.A. | | 登録日 | 2018-01-24 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | An NAD+Phosphorylase Toxin Triggers Mycobacterium tuberculosis Cell Death.
Mol.Cell, 73, 2019
|
|
8G2W
 
 | | Cryo-EM structure of 3DVA component 2 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | 分子名称: | DNA (31-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2023-02-06 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G4W
 
 | | Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | 分子名称: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-mer), ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2023-02-10 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G8Z
 
 | | Cryo-EM structure of 3DVA component 1 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | 分子名称: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-MER), ... | | 著者 | Porta, J.C, Ohi, M.D, Walter, N.G, Frank, A.T, Deb, I, Meze, K. | | 登録日 | 2023-02-20 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3UNE
 
 | | Mouse constitutive 20S proteasome | | 分子名称: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | 著者 | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | 登録日 | 2011-11-15 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
3ULF
 
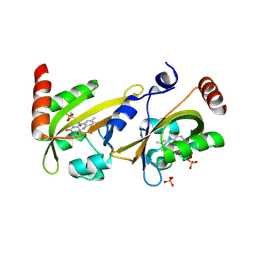 | |
6FV3
 
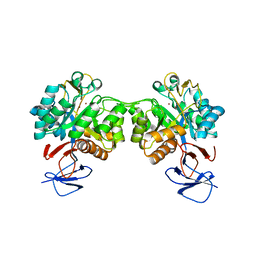 | | Crystal structure of N-acetyl-D-glucosamine-6-phosphate deacetylase from Mycobacterium smegmatis. | | 分子名称: | N-acetylglucosamine-6-phosphate deacetylase, ZINC ION | | 著者 | Ahangar, M.S, Furze, C.M, Guy, C.S, Cooper, C, Maskew, K.S, Graham, B, Cameron, A.D, Fullam, E. | | 登録日 | 2018-02-28 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structural and functional determination of homologs of theMycobacterium tuberculosis N-acetylglucosamine-6-phosphate deacetylase (NagA).
J. Biol. Chem., 293, 2018
|
|
8G7E
 
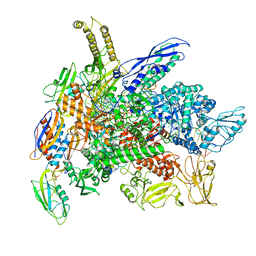 | | Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | 分子名称: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-mer), ... | | 著者 | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | 登録日 | 2023-02-16 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8GK3
 
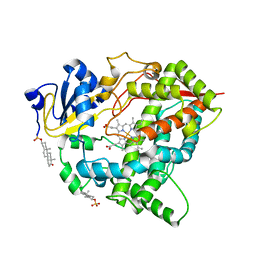 | |
7FEQ
 
 | | Cryo-EM structure of apo BsClpP at pH 6.5 | | 分子名称: | ATP-dependent Clp protease proteolytic subunit | | 著者 | Kim, L, Lee, B.-G, Kim, M.K, Kwon, D.H, Kim, H, Brotz-Oesterhelt, H, Roh, S.-H, Song, H.K. | | 登録日 | 2021-07-21 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
6FLQ
 
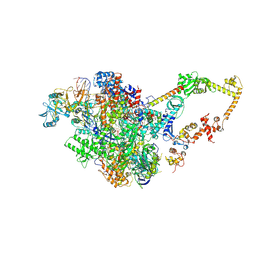 | |
8GU0
 
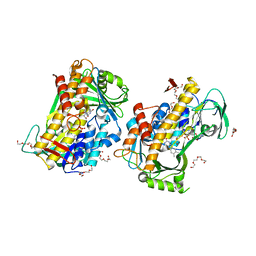 | |
3UE6
 
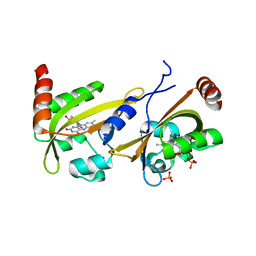 | |
6G8M
 
 | | Yeast 20S proteasome in complex with Cystargolide B Derivative 1 | | 分子名称: | (2~{S},3~{R})-4-[[(2~{S})-3-methyl-1-[[(2~{S})-3-methyl-1-oxidanylidene-1-phenylmethoxy-butan-2-yl]amino]-1-oxidanylidene-butan-2-yl]amino]-3-oxidanyl-4-oxidanylidene-2-propan-2-yl-butanoic acid, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Groll, M, Tello-Aburto, R. | | 登録日 | 2018-04-09 | | 公開日 | 2018-09-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Design, synthesis, and evaluation of cystargolide-based beta-lactones as potent proteasome inhibitors.
Eur J Med Chem, 157, 2018
|
|
8FFZ
 
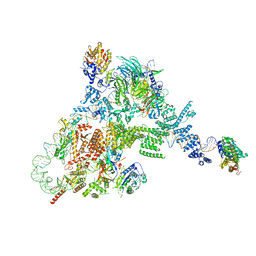 | | TFIIIA-TFIIIC-Brf1-TBP complex bound to 5S rRNA gene | | 分子名称: | DNA (151-MER), Transcription factor IIIA, Transcription factor IIIB 70 kDa subunit,TATA-box-binding protein, ... | | 著者 | Talyzina, A, He, Y. | | 登録日 | 2022-12-11 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of TFIIIC-dependent RNA polymerase III transcription initiation.
Mol.Cell, 83, 2023
|
|
4EJJ
 
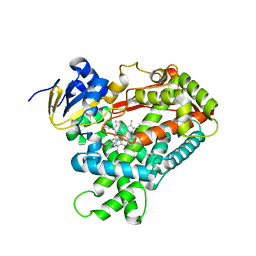 | | Human Cytochrome P450 2A6 in complex with nicotine | | 分子名称: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | DeVore, N.M, Scott, E.E. | | 登録日 | 2012-04-06 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone binding and access channel in human cytochrome P450 2A6 and 2A13 enzymes.
J.Biol.Chem., 287, 2012
|
|
3UNH
 
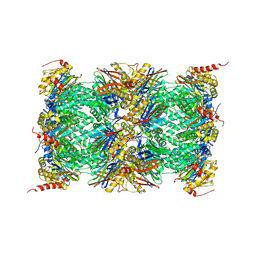 | | Mouse 20S immunoproteasome | | 分子名称: | CHLORIDE ION, IODIDE ION, POTASSIUM ION, ... | | 著者 | Huber, E, Basler, M, Schwab, R, Heinemeyer, W, Kirk, C, Groettrup, M, Groll, M. | | 登録日 | 2011-11-15 | | 公開日 | 2012-02-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity.
Cell(Cambridge,Mass.), 148, 2012
|
|
7FES
 
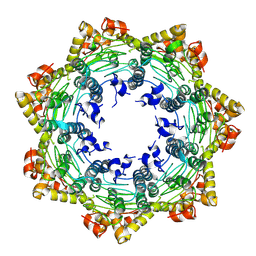 | | Cryo-EM structure of apo BsClpP at pH 4.2 | | 分子名称: | ATP-dependent Clp protease proteolytic subunit | | 著者 | Kim, L, Lee, B.-G, Kim, M.K, Kwon, D.H, Kim, H, Brotz-Oesterhelt, H, Roh, S.-H, Song, H.K. | | 登録日 | 2021-07-21 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
