7EYF
 
 | |
7EYE
 
 | |
7EYQ
 
 | |
7V0V
 
 | | GFP Nanobody NMR Structure | | 分子名称: | Anti-GFP Nanobody | | 著者 | Mueller, G.A. | | 登録日 | 2022-05-11 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nanobody Paratope Ensembles in Solution Characterized by MD Simulations and NMR.
Int J Mol Sci, 23, 2022
|
|
7QIX
 
 | | Specific features and methylation sites of a plant ribosome. 40S body ribosomal subunit. | | 分子名称: | 18S rRNA body, 30S ribosomal protein S15, chloroplastic, ... | | 著者 | Cottilli, P, Itoh, Y, Amunts, A. | | 登録日 | 2021-12-16 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.53 Å) | | 主引用文献 | Cryo-EM structure and rRNA modification sites of a plant ribosome.
Plant Commun., 3, 2022
|
|
7SSX
 
 | | Structure of human Kv1.3 | | 分子名称: | POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, Green fluorescent protein fusion | | 著者 | Meyerson, J.R, Selvakumar, P. | | 登録日 | 2021-11-11 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
7SSZ
 
 | |
7ZHG
 
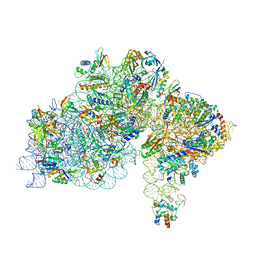 | | High-resolution cryo-EM structure of Pyrococcus abyssi 30S ribosomal subunit bound to mRNA and initiator tRNA anticodon stem-loop | | 分子名称: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | 著者 | Kazan, R, Bourgeois, G, Mechulam, Y, Coureux, P.D, Schmitt, E. | | 登録日 | 2022-04-06 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.25 Å) | | 主引用文献 | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
7SSY
 
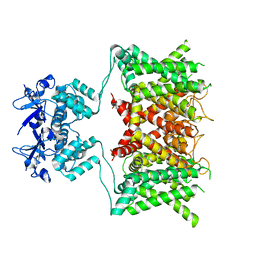 | |
7SSV
 
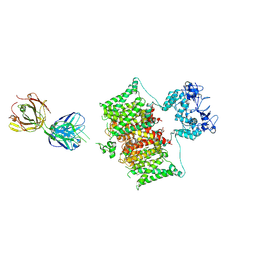 | | Structure of human Kv1.3 with Fab-ShK fusion | | 分子名称: | Fab-ShK fusion, heavy chain, light chain, ... | | 著者 | Meyerson, J.R, Selvakumar, P, Smider, V, Huang, R. | | 登録日 | 2021-11-11 | | 公開日 | 2022-06-29 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Structures of the T cell potassium channel Kv1.3 with immunoglobulin modulators.
Nat Commun, 13, 2022
|
|
7XB0
 
 | | Crystal structure of Omicron BA.2 RBD complexed with hACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Li, L, Liao, H, Meng, Y, Li, W. | | 登録日 | 2022-03-19 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7XAZ
 
 | | Crystal structure of Omicron BA.1.1 RBD complexed with hACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Liao, H, Meng, Y, Li, W. | | 登録日 | 2022-03-19 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7XB1
 
 | | Crystal structure of Omicron BA.3 RBD complexed with hACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Li, W, Meng, Y, Liao, H. | | 登録日 | 2022-03-19 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7SUK
 
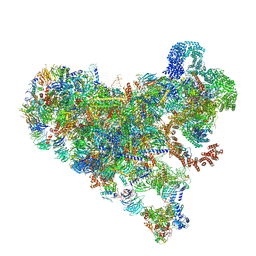 | | Structure of Bfr2-Lcp5 Complex Observed in the Small Subunit Processome Isolated from R2TP-depleted Yeast Cells | | 分子名称: | 18S pre-rRNA, 40S ribosomal protein S11-A, 40S ribosomal protein S13, ... | | 著者 | Rai, J, Zhao, Y, Li, H. | | 登録日 | 2021-11-17 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.99 Å) | | 主引用文献 | Artificial intelligence-assisted cryoEM structure of Bfr2-Lcp5 complex observed in the yeast small subunit processome.
Commun Biol, 5, 2022
|
|
7ZC2
 
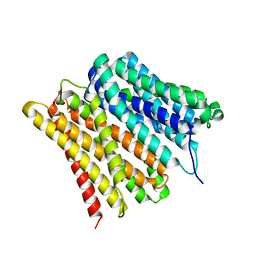 | | Dipeptide and tripeptide Permease C (DtpC) | | 分子名称: | Amino acid/peptide transporter | | 著者 | Killer, M, Finocchio, G, Pardon, E, Steyaert, J, Loew, C. | | 登録日 | 2022-03-25 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | Cryo-EM Structure of an Atypical Proton-Coupled Peptide Transporter: Di- and Tripeptide Permease C.
Front Mol Biosci, 9, 2022
|
|
8DFL
 
 | |
7OYA
 
 | | Cryo-EM structure of the 1 hpf zebrafish embryo 80S ribosome | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | 登録日 | 2021-06-24 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-02-08 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
7OYB
 
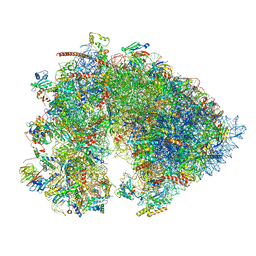 | | Cryo-EM structure of the 6 hpf zebrafish embryo 80S ribosome | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | 登録日 | 2021-06-24 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
7SYN
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 8(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYJ
 
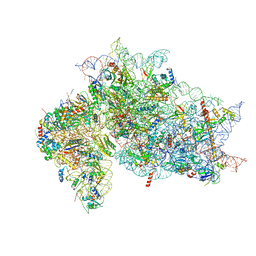 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 4(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYL
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, closed conformation. Structure 6(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYX
 
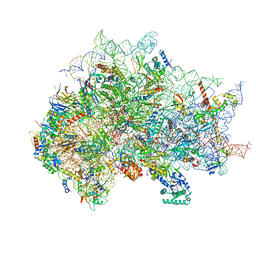 | | Structure of the delta dII IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S24, 40S ribosomal protein S25, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-03-08 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYW
 
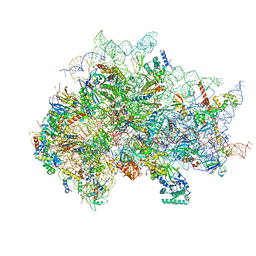 | | Structure of the wt IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(wt) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYG
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 1(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYI
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 3(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
