7N3C
 
 | | Crystal Structure of Human Fab S24-202 in the complex with the N-terminal Domain of Nucleocapsid protein from SARS CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, IODIDE ION, Nucleoprotein, ... | | 著者 | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-05-31 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7U2J
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Gly-NH-tRNAgly, peptidyl P-site fMAC-NH-tRNAmet, deacylated E-site tRNAgly, and chloramphenicol at 2.55A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | 登録日 | 2022-02-24 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural basis for the inability of chloramphenicol to inhibit peptide bond formation in the presence of A-site glycine.
Nucleic Acids Res., 50, 2022
|
|
7N3D
 
 | | Crystal Structure of Human Fab S24-1564 in the complex with the N-terminal Domain of Nucleocapsid protein from SARS CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Nucleoprotein, ... | | 著者 | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-05-31 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7NB0
 
 | |
7U2I
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Gly-NH-tRNAgly, aminoacylated P-site fMet-NH-tRNAmet, deacylated E-site tRNAgly, and chloramphenicol at 2.55A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | 登録日 | 2022-02-24 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural basis for the inability of chloramphenicol to inhibit peptide bond formation in the presence of A-site glycine.
Nucleic Acids Res., 50, 2022
|
|
8X1A
 
 | | Crystal structure of periplasmic G6P binding protein VcA0625 | | 分子名称: | 6-O-phosphono-alpha-D-glucopyranose, Iron(III) ABC transporter, periplasmic iron-compound-binding protein | | 著者 | Dasgupta, J, Saha, I. | | 登録日 | 2023-11-06 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.604 Å) | | 主引用文献 | Structural insights in to the atypical type-I ABC Glucose-6-phosphate importer VCA0625-27 of Vibrio cholerae.
Biochem.Biophys.Res.Commun., 716, 2024
|
|
7MWY
 
 | |
7MWZ
 
 | |
3FRL
 
 | | The 2.25 A crystal structure of LipL32, the major surface antigen of Leptospira interrogans serovar Copenhageni | | 分子名称: | 2,2',2''-NITRILOTRIETHANOL, CHLORIDE ION, LipL32, ... | | 著者 | Farah, C.S, Guzzo, C.R, Hauk, P, Ho, P.L. | | 登録日 | 2009-01-08 | | 公開日 | 2009-06-16 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure and calcium-binding activity of LipL32, the major surface antigen of pathogenic Leptospira sp.
J.Mol.Biol., 390, 2009
|
|
4EOY
 
 | |
8W2L
 
 | | TRPM7 structure in complex with anticancer agent CCT128930 in closed state | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-aminium, ... | | 著者 | Nadezhdin, K.D, Sobolevsky, A.I. | | 登録日 | 2024-02-20 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.45 Å) | | 主引用文献 | Structural basis of selective TRPM7 inhibition by the anticancer agent CCT128930.
Cell Rep, 43, 2024
|
|
7NUU
 
 | | Crystal structure of human AMDHD2 in complex with Zn | | 分子名称: | GLYCEROL, N-acetylglucosamine-6-phosphate deacetylase, ZINC ION | | 著者 | Ruegenberg, S, Kroef, V, Baumann, U, Denzel, M.S. | | 登録日 | 2021-03-14 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.836 Å) | | 主引用文献 | GFPT2/GFAT2 and AMDHD2 act in tandem to control the hexosamine pathway.
Elife, 11, 2022
|
|
7NUT
 
 | | Crystal structure of human AMDHD2 in complex with Zn and GlcN6P | | 分子名称: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, N-acetylglucosamine-6-phosphate deacetylase, ZINC ION | | 著者 | Ruegenberg, S, Kroef, V, Baumann, U, Denzel, M.S. | | 登録日 | 2021-03-14 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.898 Å) | | 主引用文献 | GFPT2/GFAT2 and AMDHD2 act in tandem to control the hexosamine pathway.
Elife, 11, 2022
|
|
7SNH
 
 | | Structure of G6PD-D200N tetramer bound to NADP+ | | 分子名称: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wei, X, Marmorstein, R. | | 登録日 | 2021-10-28 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SVQ
 
 | | Crystal Structure of L-galactose dehydrogenase from Spinacia oleracea in complex with NAD+ | | 分子名称: | L-galactose dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | 登録日 | 2021-11-19 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Characterization of L-Galactose Dehydrogenase: An Essential Enzyme for Vitamin C Biosynthesis.
Plant Cell.Physiol., 63, 2022
|
|
7SNF
 
 | | Structure of G6PD-WT dimer | | 分子名称: | Glucose-6-phosphate 1-dehydrogenase | | 著者 | Wei, X, Marmorstein, R. | | 登録日 | 2021-10-28 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SMI
 
 | | Crystal Structure of L-galactose dehydrogenase from Spinacia oleracea | | 分子名称: | L-galactose dehydrogenase | | 著者 | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | 登録日 | 2021-10-26 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural Characterization of L-Galactose Dehydrogenase: An Essential Enzyme for Vitamin C Biosynthesis.
Plant Cell.Physiol., 63, 2022
|
|
7SNM
 
 | |
7T64
 
 | | Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedded in lipidic nanodisc in the closed state | | 分子名称: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | 著者 | Iyer, K.A, Hu, Y, Murayama, T, Samso, M. | | 登録日 | 2021-12-13 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Molecular mechanism of the severe MH/CCD mutation Y522S in skeletal ryanodine receptor (RyR1) by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7T65
 
 | | Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedded in lipidic nanodisc in the open state | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | 著者 | Iyer, K.A, Hu, Y, Murayama, T, Samso, M. | | 登録日 | 2021-12-13 | | 公開日 | 2022-07-20 | | 最終更新日 | 2022-08-03 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Molecular mechanism of the severe MH/CCD mutation Y522S in skeletal ryanodine receptor (RyR1) by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8UM2
 
 | |
8UM1
 
 | |
7OOP
 
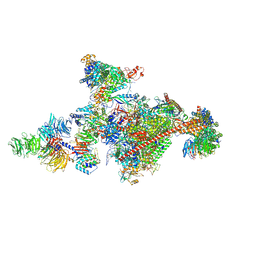 | | Pol II-CSB-CSA-DDB1-UVSSA-PAF-SPT6 (Structure 3) | | 分子名称: | DNA damage-binding protein 1, DNA excision repair protein ERCC-6, DNA excision repair protein ERCC-8, ... | | 著者 | Kokic, G, Cramer, P. | | 登録日 | 2021-05-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of human transcription-DNA repair coupling.
Nature, 598, 2021
|
|
8U4F
 
 | |
8U4A
 
 | |
