4U9I
 
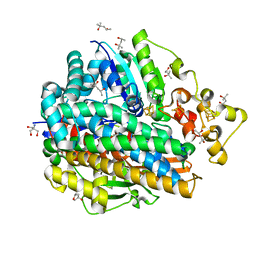 | | High Resolution Structure Of The Ni-R State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | 著者 | Ogata, H, Nishikawa, K, Lubitz, W. | | 登録日 | 2014-08-06 | | 公開日 | 2015-01-21 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | Hydrogens detected by subatomic resolution protein crystallography in a [NiFe] hydrogenase.
Nature, 520, 2015
|
|
4UDF
 
 | | STRUCTURAL BASIS OF HUMAN PARECHOVIRUS NEUTRALIZATION BY HUMAN MONOCLONAL ANTIBODIES | | 分子名称: | Capsid protein VP0, Capsid protein VP3, HUMAN MONOCLONAL ANTIBODY | | 著者 | Shakeel, S, Westerhuis, B.M, Ora, A, Koen, G, Bakker, A.Q, Claassen, Y, Beaumont, T, Wolthers, K.C, Butcher, S.J. | | 登録日 | 2014-12-10 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (20 Å) | | 主引用文献 | Structural Basis of Human Parechovirus Neutralization by Human Monoclonal Antibodies.
J. Virol., 89, 2015
|
|
4TVS
 
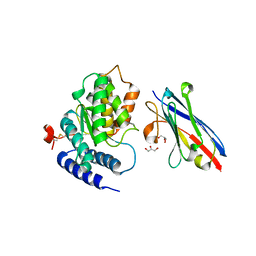 | | LAP1(aa356-583), H.sapiens, bound to VHH-BS1 | | 分子名称: | GLYCEROL, SULFATE ION, Torsin-1A-interacting protein 1, ... | | 著者 | Sosa, B.A, Schwartz, T.U. | | 登録日 | 2014-06-27 | | 公開日 | 2014-09-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | How lamina-associated polypeptide 1 (LAP1) activates Torsin.
Elife, 3, 2014
|
|
4TWB
 
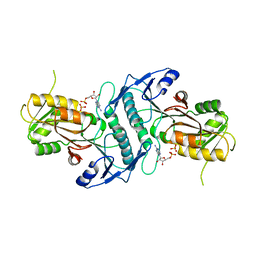 | | Sulfolobus solfataricus ribose-phosphate pyrophosphokinase | | 分子名称: | ADENOSINE MONOPHOSPHATE, Ribose-phosphate pyrophosphokinase, SULFATE ION | | 著者 | Kadziola, A. | | 登録日 | 2014-06-30 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.802 Å) | | 主引用文献 | Structure of dimeric, recombinant Sulfolobus solfataricus phosphoribosyl diphosphate synthase: a bent dimer defining the adenine specificity of the substrate ATP.
Extremophiles, 19, 2015
|
|
4TWM
 
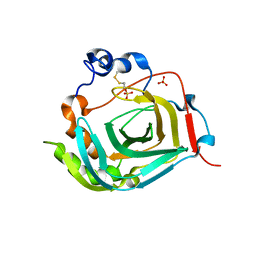 | | Crystal structure of dioscorin from Dioscorea japonica | | 分子名称: | Dioscorin 5, SULFATE ION | | 著者 | Xue, Y.L, Miyakawa, T, Nakamura, A, Tanokura, M. | | 登録日 | 2014-07-01 | | 公開日 | 2015-04-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Yam Tuber Storage Protein Reduces Plant Oxidants Using the Coupled Reactions as Carbonic Anhydrase and Dehydroascorbate Reductase
Mol Plant, 8, 2015
|
|
4TWR
 
 | | Structure of UDP-glucose 4-epimerase from Brucella abortus | | 分子名称: | NAD binding site:NAD-dependent epimerase/dehydratase:UDP-glucose 4-epimerase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | 著者 | Horanyi, P.S, Abendroth, J, Lorimer, D, Edwards, T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2014-07-01 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of UDP-glucose 4-epimerase from Brucella melitensis
To Be Published
|
|
4U95
 
 | | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB | | 分子名称: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, DARPin, DODECYL-BETA-D-MALTOSIDE, ... | | 著者 | Pos, K.M. | | 登録日 | 2014-08-05 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB
eLife, 3, 2014
|
|
4U88
 
 | |
4U8H
 
 | | Crystal Structure of Mammalian Period-Cryptochrome Complex | | 分子名称: | Cryptochrome-2, Period circadian protein homolog 2, ZINC ION | | 著者 | Nangle, S.N, Rosensweig, C, Koike, N, Tei, H, Takahashi, J.S, Green, C.B, Zheng, N. | | 登録日 | 2014-08-03 | | 公開日 | 2014-10-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.798 Å) | | 主引用文献 | Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex.
Elife, 3, 2014
|
|
4UA9
 
 | |
4UBO
 
 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 3.70 MGy TEMP 150K | | 分子名称: | DIETHYL HYDROGEN PHOSPHATE, E3 | | 著者 | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | 登録日 | 2014-08-13 | | 公開日 | 2015-08-19 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
4UE3
 
 | | The Mechanism of Hydrogen Activation by NiFe-hydrogenases and the Importance of the active site Arginine | | 分子名称: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | 著者 | Evans, R.M, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | 登録日 | 2014-12-15 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Mechanism of Hydrogen Activation by [Nife] Hydrogenases.
Nat.Chem.Biol., 12, 2016
|
|
4UDY
 
 | | NCO- bound to cluster C of Ni,Fe-CO dehydrogenase at true-atomic resolution | | 分子名称: | CARBON MONOXIDE DEHYDROGENASE 2, FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | 著者 | Fesseler, J, Jeoung, J.-H, Dobbek, H. | | 登録日 | 2014-12-12 | | 公開日 | 2015-05-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | How the [Nife4 S4 ] Cluster of Co Dehydrogenase Activates Co2 and Nco(.)
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4U46
 
 | | Crystal structure of an avidin mutant | | 分子名称: | Avidin, CHLORIDE ION | | 著者 | Agrawal, N, Lehtonen, S, Kahkonen, N, Riihimaki, T, Hytonen, V.P, Kulomaa, M.S, Johnson, M.S, Airenne, T.T. | | 登録日 | 2014-07-23 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Artificial Avidin-Based Receptors for a Panel of Small Molecules.
Acs Chem.Biol., 11, 2016
|
|
4U4C
 
 | | The molecular architecture of the TRAMP complex reveals the organization and interplay of its two catalytic activities | | 分子名称: | 1,2-ETHANEDIOL, ATP-dependent RNA helicase DOB1, CHLORIDE ION, ... | | 著者 | Falk, S, Weir, J.R, Hentschel, J, Reichelt, P, Bonneau, F, Conti, E. | | 登録日 | 2014-07-23 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The Molecular Architecture of the TRAMP Complex Reveals the Organization and Interplay of Its Two Catalytic Activities.
Mol.Cell, 55, 2014
|
|
4U4I
 
 | | Megavirus chilensis superoxide dismutase | | 分子名称: | Cu/Zn superoxide dismutase | | 著者 | Lartigue, A, Claverie, J.-M, Burlat, B, Coutard, B, Abergel, C. | | 登録日 | 2014-07-23 | | 公開日 | 2014-11-05 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The megavirus chilensis cu,zn-superoxide dismutase: the first viral structure of a typical cellular copper chaperone-independent hyperstable dimeric enzyme.
J.Virol., 89, 2015
|
|
4U3J
 
 | | TOG2:alpha/beta-tubulin complex | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein STU2, ... | | 著者 | Ayaz, P, Rice, L.M. | | 登録日 | 2014-07-22 | | 公開日 | 2014-08-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | A tethered delivery mechanism explains the catalytic action of a microtubule polymerase.
Elife, 3, 2014
|
|
4U3T
 
 | |
4U5J
 
 | | C-Src in complex with Ruxolitinib | | 分子名称: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Proto-oncogene tyrosine-protein kinase Src | | 著者 | Chen, Y, Duan, Y, Chen, L. | | 登録日 | 2014-07-25 | | 公開日 | 2014-09-17 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | c-Src Binds to the Cancer Drug Ruxolitinib with an Active Conformation
Plos One, 9, 2014
|
|
4U5Q
 
 | | High resolution crystal structure of reductase (R) domain of nonribosomal peptide synthetase from Mycobacterium tuberculosis | | 分子名称: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Peptide synthetase | | 著者 | Patel, K.D, Haque, A.S, Priyadarshan, K, Sankaranarayanan, R. | | 登録日 | 2014-07-25 | | 公開日 | 2016-01-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.811 Å) | | 主引用文献 | High resolution structure of R-domain of nonribosomal peptide synthetase from Mycobacterium tuberculosis
To Be Published
|
|
4U5Y
 
 | | Crystal Structure of the complex between the GNAT domain of S. lividans PAT and the acetyl-CoA synthetase C-terminal domain of S. enterica | | 分子名称: | Acetyl-coenzyme A synthetase, Acyl-CoA synthetase, SULFATE ION | | 著者 | Taylor, K.C, Tucker, A.C, Rank, K.C, Escalante-Semerena, J.C, Rayment, I. | | 登録日 | 2014-07-25 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.926 Å) | | 主引用文献 | Insights into the specificity of lysine acetyltransferases.
J.Biol.Chem., 289, 2014
|
|
4U68
 
 | |
4U6E
 
 | | HsMetAP in complex with (amino(phenyl)methyl)phosphonic acid | | 分子名称: | COBALT (II) ION, Methionine aminopeptidase 1, [(R)-amino(phenyl)methyl]phosphonic acid | | 著者 | Arya, T, Addlagatta, A. | | 登録日 | 2014-07-28 | | 公開日 | 2015-03-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Identification of the Molecular Basis of Inhibitor Selectivity between the Human and Streptococcal Type I Methionine Aminopeptidases
J.Med.Chem., 58, 2015
|
|
4U6Z
 
 | |
4U87
 
 | | Crystal structure of the Ba-soaked C2 crystal form of pMV158 replication initiator RepB (P3221 space group) | | 分子名称: | BARIUM ION, CHLORIDE ION, MANGANESE (II) ION, ... | | 著者 | Boer, D.R, Ruiz Maso, J.A, del Solar, G, Coll, M. | | 登録日 | 2014-08-01 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Conformational plasticity of RepB, the replication initiator protein of promiscuous streptococcal plasmid pMV158.
Sci Rep, 6, 2016
|
|
