3OJO
 
 | | Derivative structure of the UDP-N-acetyl-mannosamine dehydrogenase Cap5O from S. aureus | | 分子名称: | Cap5O, EUROPIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Nessler, S, Gruszczyk, J, Olivares-Illana, V, Meyer, P, Morera, S. | | 登録日 | 2010-08-23 | | 公開日 | 2011-03-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure Analysis of the Staphylococcus aureus UDP-N-acetyl-mannosamine Dehydrogenase Cap5O Involved in Capsular Polysaccharide Biosynthesis.
J.Biol.Chem., 286, 2011
|
|
1BDB
 
 | | CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE FROM PSEUDOMONAS SP. LB400 | | 分子名称: | CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Huelsmeyer, M, Hecht, H.-J, Niefind, K, Hofer, B, Timmis, K.N, Schomburg, D. | | 登録日 | 1997-05-10 | | 公開日 | 1997-11-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of cis-biphenyl-2,3-dihydrodiol-2,3-dehydrogenase from a PCB degrader at 2.0 A resolution.
Protein Sci., 7, 1998
|
|
4J1O
 
 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound l-proline betaine (substrate) | | 分子名称: | 1,1-DIMETHYL-PROLINIUM, GLYCEROL, IODIDE ION, ... | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, LaFleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-02-01 | | 公開日 | 2013-03-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
1UP4
 
 | |
1V4B
 
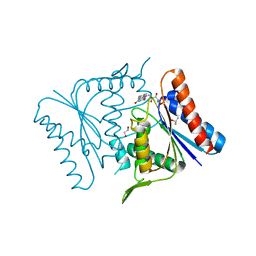 | |
3R3S
 
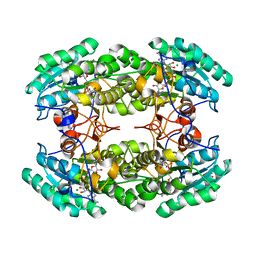 | | Structure of the YghA Oxidoreductase from Salmonella enterica | | 分子名称: | FORMIC ACID, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-03-16 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Structure of the YghA Oxidoreductase from Salmonella enterica
TO BE PUBLISHED
|
|
4EOT
 
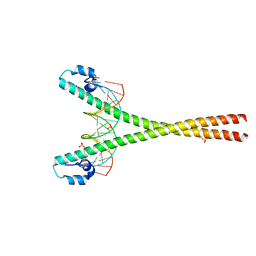 | | Crystal structure of the MafA homodimer bound to the consensus MARE | | 分子名称: | 5'-D(*CP*CP*CP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*GP*CP*AP*CP*CP*G)-3', 5'-D(*CP*CP*GP*GP*TP*GP*CP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*GP*G)-3', SULFATE ION, ... | | 著者 | Lu, X, Guanga, G, Wan, C, Rose, R.B. | | 登録日 | 2012-04-15 | | 公開日 | 2012-11-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.855 Å) | | 主引用文献 | A Novel DNA Binding Mechanism for maf Basic Region-Leucine Zipper Factors Inferred from a MafA-DNA Complex Structure and Binding Specificities.
Biochemistry, 51, 2012
|
|
1UP6
 
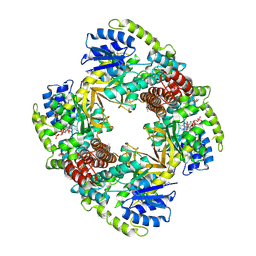 | | Structure of the 6-phospho-beta glucosidase from Thermotoga maritima at 2.55 Angstrom resolution in the tetragonal form with manganese, NAD+ and glucose-6-phosphate | | 分子名称: | 6-O-phosphono-alpha-D-glucopyranose, 6-PHOSPHO-BETA-GLUCOSIDASE, MANGANESE (II) ION, ... | | 著者 | Varrot, A, Yip, V.L, Withers, S.G, Davies, G.J. | | 登録日 | 2003-09-29 | | 公開日 | 2004-08-02 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | An Unusual Mechanism of Glycoside Hydrolysis Involving Redox and Elimination Steps by a Family 4 Beta-Glycosidase from Thermotoga Maritima.
J.Am.Chem.Soc., 126, 2004
|
|
3SQ8
 
 | |
3OUT
 
 | | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate. | | 分子名称: | D-GLUTAMIC ACID, Glutamate racemase | | 著者 | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Kudriska, M, Edwards, A, Savchenko, A, Anderson, F.W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-09-15 | | 公開日 | 2010-09-29 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate.
To be Published
|
|
4G47
 
 | | Structure of cytochrome P450 CYP121 in complex with 4-(1H-1,2,4-triazol-1-yl)phenol | | 分子名称: | 4-(1H-1,2,4-triazol-1-yl)phenol, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Hudson, S.A, McLean, K.J, Surade, S, Yang, Y.-Q, Leys, D, Ciulli, A, Munro, A.W, Abell, C. | | 登録日 | 2012-07-16 | | 公開日 | 2012-09-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4G44
 
 | | Structure of P450 CYP121 in complex with lead compound MB286, 3-((1H-1,2,4-triazol-1-yl)methyl)aniline | | 分子名称: | 3-(1H-1,2,4-triazol-1-ylmethyl)aniline, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Hudson, S.A, McLean, K.J, Surade, S, Yang, Y.-Q, Leys, D, Ciulli, A, Munro, A.W, Abell, C. | | 登録日 | 2012-07-16 | | 公開日 | 2012-09-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3T3Q
 
 | | Human Cytochrome P450 2A6 I208S/I300F/G301A/S369G in complex with Pilocarpine | | 分子名称: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | DeVore, N.M, Scott, E.E. | | 登録日 | 2011-07-25 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
3SQ7
 
 | |
3UAS
 
 | | Cytochrome P450 2B4 covalently bound to the mechanism-based inactivator 9-ethynylphenanthrene | | 分子名称: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Gay, S.C, Zhang, H, Shah, M.B, Stout, C.D, Halpert, J.R, Hollenberg, P.F. | | 登録日 | 2011-10-21 | | 公開日 | 2013-01-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.939 Å) | | 主引用文献 | Potent Mechanism-Based Inactivation of Cytochrome P450 2B4 by 9-Ethynylphenanthrene: Implications for Allosteric Modulation of Cytochrome P450 Catalysis.
Biochemistry, 52, 2013
|
|
4IZG
 
 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound cis-4oh-d-proline betaine (product) | | 分子名称: | (2S,4S)-2-carboxy-4-hydroxy-1,1-dimethylpyrrolidinium, IODIDE ION, MAGNESIUM ION, ... | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, LaFleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-01-29 | | 公開日 | 2013-02-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
4IS2
 
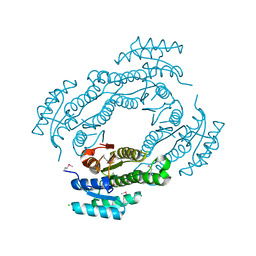 | |
4GQS
 
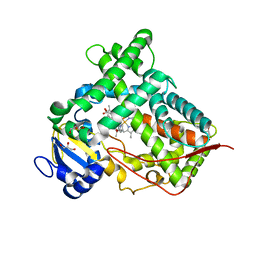 | | Structure of Human Microsomal Cytochrome P450 (CYP) 2C19 | | 分子名称: | (4-hydroxy-3,5-dimethylphenyl)(2-methyl-1-benzofuran-3-yl)methanone, Cytochrome P450 2C19, GLYCEROL, ... | | 著者 | Reynald, R.L, Sansen, S, Stout, C.D, Johnson, E.F. | | 登録日 | 2012-08-23 | | 公開日 | 2012-11-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Structural Characterization of Human Cytochrome P450 2C19: ACTIVE SITE DIFFERENCES BETWEEN P450s 2C8, 2C9, AND 2C19.
J.Biol.Chem., 287, 2012
|
|
3S81
 
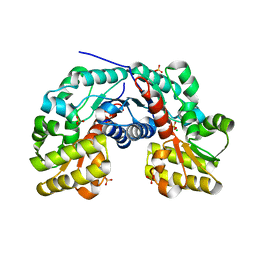 | | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium | | 分子名称: | CHLORIDE ION, Putative aspartate racemase, SULFATE ION | | 著者 | Maltseva, N, Kim, Y, Kwon, K, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-05-27 | | 公開日 | 2011-06-15 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.796 Å) | | 主引用文献 | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium
To be Published
|
|
1UT5
 
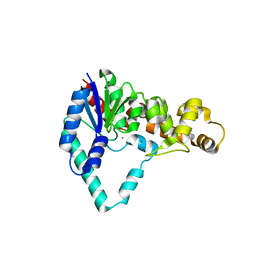 | |
1UT8
 
 | |
3LT5
 
 | | X-ray Crystallographic structure of a Pseudomonas Aeruginosa Azoreductase in complex with balsalazide | | 分子名称: | (3E)-3-({4-[(2-carboxyethyl)carbamoyl]phenyl}hydrazono)-6-oxocyclohexa-1,4-diene-1-carboxylic acid, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, ... | | 著者 | Ryan, A, Laurieri, N, Westwood, I, Wang, C.-J, Lowe, E, Sim, E. | | 登録日 | 2010-02-15 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A Novel Mechanism for Azoreduction
J.Mol.Biol., 400, 2010
|
|
3LNP
 
 | | Crystal Structure of Amidohydrolase family Protein OLEI01672_1_465 from Oleispira antarctica | | 分子名称: | ACETIC ACID, Amidohydrolase family Protein OLEI01672_1_465, CALCIUM ION, ... | | 著者 | Kim, Y, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-02-02 | | 公開日 | 2010-02-16 | | 最終更新日 | 2013-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3U7I
 
 | | The crystal structure of FMN-dependent NADH-azoreductase 1 (GBAA0966) from Bacillus anthracis str. Ames Ancestor | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FMN-dependent NADH-azoreductase 1, ... | | 著者 | Zhang, R, Gu, M, Tan, K, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-10-13 | | 公開日 | 2011-11-09 | | 最終更新日 | 2015-12-16 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The crystal structure of FMN-dependent NADH-azoreductase 1 (GBAA0966) from Bacillus anthracis str. Ames Ancestor
To be Published
|
|
1VJT
 
 | |
