3PS2
 
 | | Crystal structure of the Escherichia Coli LPXC/LPC-012 complex | | 分子名称: | 4-[4-(3-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-nitroso-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2010-11-30 | | 公開日 | 2011-01-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Syntheses, structures and antibiotic activities of LpxC inhibitors based on the diacetylene scaffold.
Bioorg.Med.Chem., 19, 2011
|
|
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | 分子名称: | Pachytene checkpoint protein 2 homolog | | 著者 | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | 登録日 | 2019-12-17 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
3C82
 
 | |
8OWI
 
 | |
3PXF
 
 | | CDK2 in complex with two molecules of 8-anilino-1-naphthalene sulfonate | | 分子名称: | 1,2-ETHANEDIOL, 8-ANILINO-1-NAPHTHALENE SULFONATE, Cell division protein kinase 2 | | 著者 | Betzi, S, Alam, R, Schonbrunn, E. | | 登録日 | 2010-12-09 | | 公開日 | 2011-02-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of a Potential Allosteric Ligand Binding Site in CDK2.
Acs Chem.Biol., 6, 2011
|
|
6L3M
 
 | |
6L6O
 
 | | Crystal structure of stabilized Rab5a GTPase domain from Leishmania donovani | | 分子名称: | ACETATE ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Arora, A, Zohib, M, Biswal, B.K, Maheshwari, D, Pal, R.K. | | 登録日 | 2019-10-29 | | 公開日 | 2020-11-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the GDP-bound GTPase domain of Rab5a from Leishmania donovani.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
3C7Y
 
 | |
6L96
 
 | | Structure of PPARalpha-LBD/pemafibrate/SRC1 peptide | | 分子名称: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, Peroxisome proliferator-activated receptor alpha, SRC1 coactivator peptide | | 著者 | Kawasaki, M, Kambe, A, Yamamoto, Y, Arulmozhira, S, Ito, S, Nakagawa, Y, Tokiwa, H, Nakano, S, Shimano, H. | | 登録日 | 2019-11-08 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Elucidation of Molecular Mechanism of a Selective PPAR alpha Modulator, Pemafibrate, through Combinational Approaches of X-ray Crystallography, Thermodynamic Analysis, and First-Principle Calculations.
Int J Mol Sci, 21, 2020
|
|
3LLL
 
 | | Crystal structure of mouse pacsin2 F-BAR domain | | 分子名称: | CALCIUM ION, CHLORIDE ION, Protein kinase C and casein kinase substrate in neurons protein 2, ... | | 著者 | Rudolph, M.G. | | 登録日 | 2010-01-29 | | 公開日 | 2010-05-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | A hinge in the distal end of the PACSIN 2 F-BAR domain may contribute to membrane-curvature sensing.
J.Mol.Biol., 400, 2010
|
|
3LMV
 
 | | D-Tyr-tRNA(Tyr) Deacylase from plasmodium falciparum in complex with hepes | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-tyrosyl-tRNA(Tyr) deacylase, SULFITE ION | | 著者 | Manickam, Y, Khan, S, Bhatt, T.K, Sharma, A. | | 登録日 | 2010-02-01 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.833 Å) | | 主引用文献 | Structure of D-tyrosyl-tRNATyr deacylase using home-source Cu Kalpha and moderate-quality iodide-SAD data: structural polymorphism and HEPES-bound enzyme states
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6LIO
 
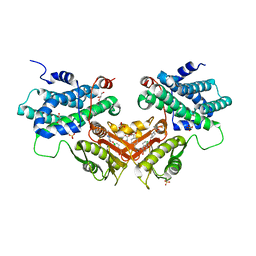 | | Crystal structure of human PDK2 complexed with GM67520 | | 分子名称: | 4-[[[5-[5-chloranyl-2,4-bis(oxidanyl)phenyl]-4-[4-(1-methylsulfonylpiperidin-4-yl)oxyphenyl]-1,2-oxazol-3-yl]carbonylamino]methyl]cyclohexane-1-carboxylic acid, GLYCEROL, SULFATE ION, ... | | 著者 | Kang, J, Kim, J. | | 登録日 | 2019-12-12 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural basis for the inhibition of PDK2 by novel ATP- and lipoyl-binding site targeting compounds.
Biochem.Biophys.Res.Commun., 527, 2020
|
|
6L1X
 
 | | Quinol-dependent nitric oxide reductase (qNOR) from Neisseria meningitidis in the monomeric oxidized state with zinc complex. | | 分子名称: | CALCIUM ION, FE (III) ION, Nitric-oxide reductase, ... | | 著者 | Jamali, M.M.A, Antonyuk, S.V, Tosha, T, Muramoto, K, Hasnain, S.S, Shiro, Y. | | 登録日 | 2019-10-01 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | The active form of quinol-dependent nitric oxide reductase fromNeisseria meningitidisis a dimer.
Iucrj, 7, 2020
|
|
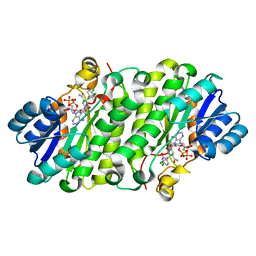
3PJE
 
 | |
6L3S
 
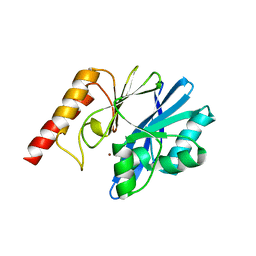 | |
6L0Q
 
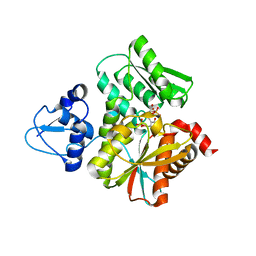 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Phosphoserine | | 分子名称: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | 著者 | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | 登録日 | 2019-09-26 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
6L25
 
 | |
6L90
 
 | | Crystal structure of ugt transferase enzyme | | 分子名称: | Glycosyltransferase, SULFATE ION | | 著者 | Li, J, Shan, N, Yang, J.G, Liu, W.D, Sun, Y.X. | | 登録日 | 2019-11-07 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Efficient O-Glycosylation of Triterpenes Enabled by Protein Engineering of Plant Glycosyltransferase UGT74AC1
Acs Catalysis, 2020
|
|
3C8Q
 
 | |
6LI0
 
 | | Crystal structure of GPR52 in complex with agonist c17 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRATE ANION, Chimera of G-protein coupled receptor 52 and Flavodoxin, ... | | 著者 | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | 登録日 | 2019-12-10 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LFF
 
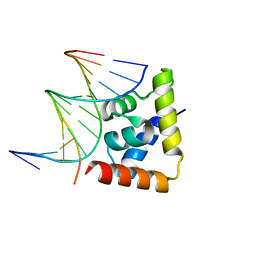 | | transcription factor SATB1 CUTr1 domain in complex with a phosphorothioate DNA | | 分子名称: | DNA (5'-D(*GP*(C7R)P*(PST)P*AP*AP*TP*AP*TP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*(AS)P*(PST)P*(AS)P*(PST)P*TP*AP*GP*C)-3'), DNA-binding protein SATB1 | | 著者 | Akutsu, Y, Kubota, T, Yamasaki, T, Yamasaki, K. | | 登録日 | 2019-12-02 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Enhanced affinity of racemic phosphorothioate DNA with transcription factor SATB1 arising from diastereomer-specific hydrogen bonds and hydrophobic contacts.
Nucleic Acids Res., 48, 2020
|
|
3CDT
 
 | |
3LS2
 
 | |
6L0P
 
 | | Crystal Structure of the O-Phosphoserine Sulfhydrylase from Aeropyrum pernix Complexed with O-Phosphoserine | | 分子名称: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Protein CysO | | 著者 | Nakabayashi, M, Takeda, E, Ishikawa, K, Nakamura, T. | | 登録日 | 2019-09-26 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Identification of amino acid residues important for recognition of O-phospho-l-serine substrates by cysteine synthase.
J.Biosci.Bioeng., 131, 2021
|
|
3CDV
 
 | |
