7S0X
 
 | |
6WAB
 
 | | Crystal structure of human galectin-4 C-terminal carbohydrate recognition domain in complex with galactose derivative | | 分子名称: | 2,6-anhydro-1,4-dideoxy-1-[4-(4-fluorophenyl)-1H-1,2,3-triazol-1-yl]-4-(4-phenyl-1H-1,2,3-triazol-1-yl)-D-glycero-L-manno-heptitol, GLYCEROL, Galectin-4 | | 著者 | Go, R.M, Kishor, C, Blanchard, H. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Crystal structure of human galectin-4 C-terminal carbohydrate recognition domain in complex with galactose derivative
To Be Published
|
|
6T6I
 
 | |
6T6E
 
 | |
1Y7N
 
 | | Solution structure of the second PDZ domain of the human neuronal adaptor X11alpha | | 分子名称: | Amyloid beta A4 precursor protein-binding family A member 1 | | 著者 | Duquesne, A.E, de Ruijter, M, Brouwer, J, Drijfhout, J.W, Nabuurs, S.B, Spronk, C.A.E.M, Vuister, G.W, Ubbink, M, Canters, G.W. | | 登録日 | 2004-12-09 | | 公開日 | 2005-11-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the second PDZ domain of the neuronal adaptor X11alpha and its interaction with the C-terminal peptide of the human copper chaperone for superoxide dismutase
J.Biomol.Nmr, 32, 2005
|
|
8P9Y
 
 | | SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | 著者 | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | 登録日 | 2023-06-06 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
8P99
 
 | | SARS-CoV-2 S-protein:D614G mutant in 1-up conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1,Spike glycoprotein | | 著者 | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | 登録日 | 2023-06-05 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
5GRN
 
 | | Crystal structure of PDGFRA in Complex with WQ-C-159 | | 分子名称: | CHLORIDE ION, N-[2-(dimethylamino)ethyl]-N-[[4-[[4-methyl-3-[(4-pyridin-3-ylpyrimidin-2-yl)amino]phenyl]carbamoyl]phenyl]methyl]pyridine-3-carboxamide, Platelet-derived growth factor receptor alpha | | 著者 | Yan, X.E, Yun, C.H. | | 登録日 | 2016-08-11 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Crystal Structure of PDGFRA in Complex with WQ-C-159
To Be Published
|
|
8J2B
 
 | |
8S4X
 
 | | Tankyrase 2 in complex with a quinazolin-4-one inhibitor | | 分子名称: | 8-[bis(oxidanyl)amino]-2-(4-~{tert}-butylphenyl)-3~{H}-quinazolin-4-one, Poly [ADP-ribose] polymerase tankyrase-2, SULFATE ION, ... | | 著者 | Bosetti, C, Lehtio, L. | | 登録日 | 2024-02-22 | | 公開日 | 2025-03-05 | | 最終更新日 | 2025-03-26 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Substitutions at the C-8 position of quinazolin-4-ones improve the potency of nicotinamide site binding tankyrase inhibitors.
Eur.J.Med.Chem., 288, 2025
|
|
8J2A
 
 | |
8J2D
 
 | |
8J2C
 
 | |
8J2E
 
 | |
5OGS
 
 | | Crystal structure of human AND-1 SepB domain | | 分子名称: | MALONATE ION, WD repeat and HMG-box DNA-binding protein 1 | | 著者 | Pellegrini, L, Simon, A.C. | | 登録日 | 2017-07-13 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.503 Å) | | 主引用文献 | The human CTF4-orthologue AND-1 interacts with DNA polymerase alpha /primase via its unique C-terminal HMG box.
Open Biol, 7, 2017
|
|
5KTX
 
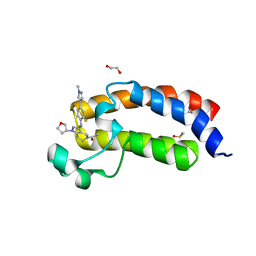 | | CREBBP bromodomain in complex with Cpd59 ((S)-1-(3-((2-fluoro-4-(1-methyl-1H-pyrazol-4-yl)phenyl)amino)-1-(tetrahydrofuran-3-yl)-6,7-dihydro-1H-pyrazolo[4,3-c]pyridin-5(4H)-yl)ethanone) | | 分子名称: | 1,2-ETHANEDIOL, 1-[3-[[2-fluoranyl-4-(1-methylpyrazol-4-yl)phenyl]amino]-1-[(3~{S})-oxolan-3-yl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-5-yl]ethanone, CREB-binding protein, ... | | 著者 | Murray, J.M, Noland, C. | | 登録日 | 2016-07-12 | | 公開日 | 2016-11-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
6FNZ
 
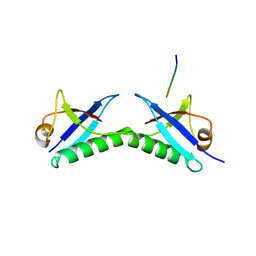 | |
8HAW
 
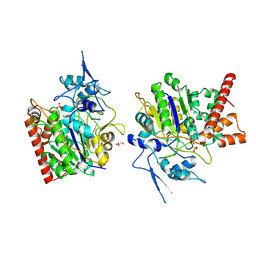 | | An auto-activation mechanism of plant non-specific phospholipase C | | 分子名称: | CALCIUM ION, GLYCEROL, Non-specific phospholipase C4, ... | | 著者 | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | 登録日 | 2022-10-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
8JN8
 
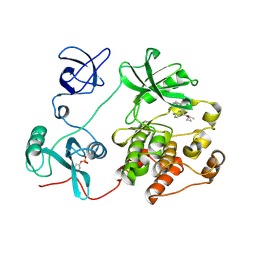 | |
8JN9
 
 | |
7B3W
 
 | | Crystal structure of c-MET bound by compound 4 | | 分子名称: | 1,2-ETHANEDIOL, 3-(6-fluoranyl-1~{H}-indazol-4-yl)-4,5-dihydro-1~{H}-pyrrolo[3,4-b]pyrrol-6-one, Hepatocyte growth factor receptor | | 著者 | Collie, G.W. | | 登録日 | 2020-12-01 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B43
 
 | | Crystal structure of c-MET bound by compound 9 | | 分子名称: | 3-[(4-fluorophenyl)methyl]-5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridine, Hepatocyte growth factor receptor | | 著者 | Collie, G.W. | | 登録日 | 2020-12-02 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
6SAZ
 
 | | Cleaved human fetuin-b in complex with crayfish astacin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Astacin, ... | | 著者 | Gomis-Ruth, F.X, Guevara, T, Cuppari, A, Korschgen, H, Schmitz, C, Kuske, M, Yiallouros, I, Floehr, J, Jahnen-Dechent, W, Stocker, W. | | 登録日 | 2019-07-18 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The C-terminal region of human plasma fetuin-B is dispensable for the raised-elephant-trunk mechanism of inhibition of astacin metallopeptidases.
Sci Rep, 9, 2019
|
|
9GFK
 
 | | human MDM2 complex with stapled foldamer | | 分子名称: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, Stapled foldamer | | 著者 | Neuville, M, Bourgeais, M, Buratto, J, Saragaglia, C, Bo, L, Mauran, L, Varajao, L, Goudreau, S.R, Kauffmann, B, Thinon, E, Pasco, M, Khatib, A.M. | | 登録日 | 2024-08-09 | | 公開日 | 2025-03-26 | | 最終更新日 | 2025-04-16 | | 実験手法 | X-RAY DIFFRACTION (1.841 Å) | | 主引用文献 | Optimal Stapling of a Helical Peptide-Foldamer Hybrid Using a C-Terminal 4-Mercaptoproline Enhances Protein Surface Recognition and Cellular Activity.
Chemistry, 31, 2025
|
|
7QCN
 
 | |
