3KN0
 
 | | Structure of BACE bound to SCH708236 | | 分子名称: | 3-[2-(3-{[(furan-2-ylmethyl)(methyl)amino]methyl}phenyl)ethyl]pyridin-2-amine, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Strickland, C, Wang, Y. | | 登録日 | 2009-11-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Application of Fragment-Based NMR Screening, X-ray Crystallography, Structure-Based Design, and Focused Chemical Library Design to Identify Novel muM Leads for the Development of nM BACE-1 (beta-Site APP Cleaving Enzyme 1) Inhibitors.
J.Med.Chem., 53, 2010
|
|
3L06
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase E92V mutant complexed with carbamyl phosphate and N-succinyl-L-norvaline | | 分子名称: | N-(3-CARBOXYPROPANOYL)-L-NORVALINE, N-acetylornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | 著者 | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | 登録日 | 2009-12-09 | | 公開日 | 2010-03-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | A single mutation in the active site swaps the substrate specificity of N-acetyl-L-ornithine transcarbamylase and N-succinyl-L-ornithine transcarbamylase.
Protein Sci., 16, 2007
|
|
3L31
 
 | | Crystal structure of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with the inhibitor, AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Probable manganase-dependent inorganic pyrophosphatase | | 著者 | Tuominen, H, Salminen, A, Oksanen, E, Jamsen, J, Heikkila, O, Lehtio, L, Magretova, N.N, Goldman, A, Baykov, A.A, Lahti, R. | | 登録日 | 2009-12-16 | | 公開日 | 2010-04-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridiumperfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate.
J.Mol.Biol., 2010
|
|
3L1E
 
 | | Bovine AlphaA crystallin Zinc Bound | | 分子名称: | Alpha-crystallin A chain, GLYCEROL, ZINC ION | | 著者 | Laganowsky, A, Sawaya, M.R, Cascio, D, Eisenberg, D. | | 登録日 | 2009-12-11 | | 公開日 | 2010-05-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function.
Protein Sci., 19, 2010
|
|
3L3V
 
 | | Structure of HIV-1 integrase core domain in complex with sucrose | | 分子名称: | CADMIUM ION, POL polyprotein, SULFATE ION, ... | | 著者 | Wielens, J, Chalmers, D.K, Scanlon, M.J, Parker, M.W. | | 登録日 | 2009-12-18 | | 公開日 | 2010-03-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the HIV-1 integrase core domain in complex with sucrose reveals details of an allosteric inhibitory binding site
Febs Lett., 584, 2010
|
|
3L4M
 
 | |
3L3U
 
 | | Crystal structure of the HIV-1 integrase core domain to 1.4A | | 分子名称: | POL polyprotein, SULFATE ION | | 著者 | Wielens, J, Chalmers, D.K, Scanlon, M.J, Parker, M.W. | | 登録日 | 2009-12-17 | | 公開日 | 2010-03-31 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of the HIV-1 integrase core domain in complex with sucrose reveals details of an allosteric inhibitory binding site.
Febs Lett., 584, 2010
|
|
3KPM
 
 | | Crystal Structure of HLA B*4402 in complex with EEYLKAWTF, a mimotope | | 分子名称: | Beta-2-microglobulin, EEYLKAWTF, mimotope peptide, ... | | 著者 | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | 登録日 | 2009-11-16 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3KQN
 
 | |
3KQU
 
 | |
3KQO
 
 | |
3KR1
 
 | |
3KS5
 
 | |
3KSD
 
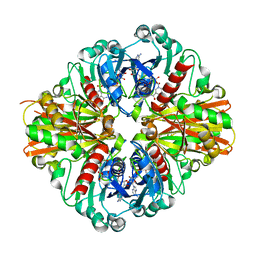 | | Crystal Structure of C151S+H178N mutant of Glyceraldehyde-3-phosphate dehydrogenase 1 (GAPDH1) from Staphylococcus aureus MRSA252 complexed with NAD at 2.2 angstrom resolution | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2009-11-22 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3KSS
 
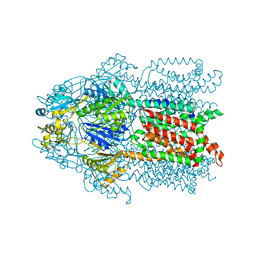 | |
3KTH
 
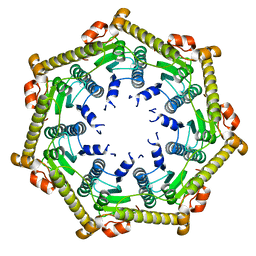 | |
3KRZ
 
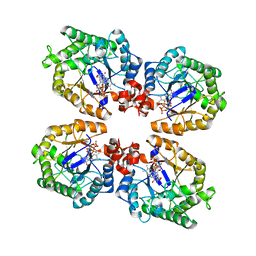 | | Crystal Structure of the Thermostable NADH4-bound old yellow enzyme from Thermoanaerobacter pseudethanolicus E39 | | 分子名称: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN MONONUCLEOTIDE, NADH:flavin oxidoreductase/NADH oxidase | | 著者 | Adalbjornsson, B.V, Toogood, H.S, Leys, D, Scrutton, N.S. | | 登録日 | 2009-11-20 | | 公開日 | 2009-12-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Biocatalysis with thermostable enzymes: structure and properties of a thermophilic 'ene'-reductase related to old yellow enzyme.
Chembiochem, 11, 2010
|
|
3KV3
 
 | | Crystal structure of C151S mutant of Glyceraldehyde-3-phosphate dehydrogenase 1 (GAPDH 1)from methicillin resistant Staphylococcus aureus MRSA252 complexed with NAD and G3P | | 分子名称: | 3-PHOSPHOGLYCERIC ACID, GAPDH, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | 登録日 | 2009-11-29 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase 1 from methicillin-resistant Staphylococcus aureus MRSA252 provides novel insights into substrate binding and catalytic mechanism.
J.Mol.Biol., 401, 2010
|
|
3KVV
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | 分子名称: | 1,4-anhydro-D-erythro-pent-1-enitol, 5-FLUOROURACIL, SULFATE ION, ... | | 著者 | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | 登録日 | 2009-11-30 | | 公開日 | 2010-04-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
3KWR
 
 | |
3KT4
 
 | | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex | | 分子名称: | FE (III) ION, PKHD-type hydroxylase TPA1 | | 著者 | Kim, H.S, Kim, H.L, Kim, K.H, Kim, D.J, Lee, S.J, Yoon, J.Y, Yoon, H.J, Lee, H.Y, Park, S.B, Kim, S.-J, Lee, J.Y, Suh, S.W. | | 登録日 | 2009-11-24 | | 公開日 | 2010-01-19 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex
Nucleic Acids Res., 38, 2010
|
|
3KTG
 
 | |
3KY8
 
 | |
3KYH
 
 | |
3KZN
 
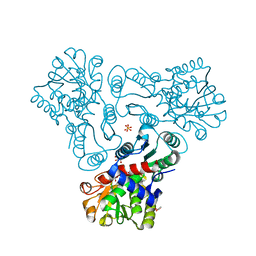 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with N-acetyl-L-ornirthine | | 分子名称: | GLYCEROL, N-acetylornithine carbamoyltransferase, N~2~-ACETYL-L-ORNITHINE, ... | | 著者 | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | 登録日 | 2009-12-08 | | 公開日 | 2010-03-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
