6GYS
 
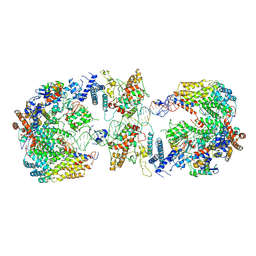 | | Cryo-EM structure of the CBF3-CEN3 complex of the budding yeast kinetochore | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-01 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6IP8
 
 | | Cryo-EM structure of the HCV IRES dependently initiated CMV-stalled 80S ribosome (Structure iv) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | 登録日 | 2018-11-02 | | 公開日 | 2019-05-29 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
6J2Q
 
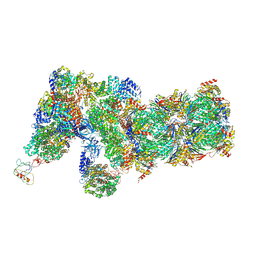 | | Yeast proteasome in Ub-accepted state (C1-b) | | 分子名称: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | 著者 | Cong, Y. | | 登録日 | 2019-01-02 | | 公開日 | 2019-03-13 | | 最終更新日 | 2019-04-10 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations.
Mol. Cell, 73, 2019
|
|
6J5J
 
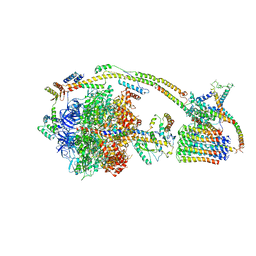 | | Cryo-EM structure of the mammalian E-state ATP synthase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F1 subunit epsilon, ... | | 著者 | Gu, J, Zhang, L, Yi, J, Yang, M. | | 登録日 | 2019-01-11 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
6HRM
 
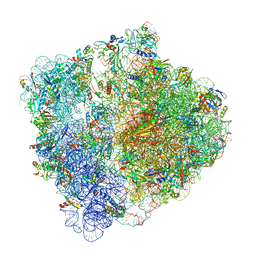 | | E. coli 70S d2d8 stapled ribosome | | 分子名称: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | 著者 | Schmied, W.H, Tnimov, Z, Uttamapinant, C, Rae, C.D, Fried, S.D, Chin, J.W. | | 登録日 | 2018-09-27 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.96 Å) | | 主引用文献 | Controlling orthogonal ribosome subunit interactions enables evolution of new function.
Nature, 564, 2018
|
|
6HUM
 
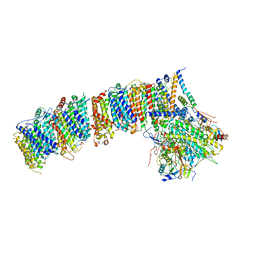 | | Structure of the photosynthetic complex I from Thermosynechococcus elongatus | | 分子名称: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, IRON/SULFUR CLUSTER, ... | | 著者 | Schuller, J.M, Schuller, S.K, Kurisu, G, Engel, B.D, Nowaczyk, M.M. | | 登録日 | 2018-10-09 | | 公開日 | 2019-01-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer.
Science, 363, 2019
|
|
6JO6
 
 | | Structure of the green algal photosystem I supercomplex with light-harvesting complex I | | 分子名称: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Suga, M, Miyazaki, N, Takahashi, Y. | | 登録日 | 2019-03-20 | | 公開日 | 2019-06-19 | | 最終更新日 | 2019-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure of the green algal photosystem I supercomplex with a decameric light-harvesting complex I.
Nat.Plants, 5, 2019
|
|
6JO5
 
 | | Structure of the green algal photosystem I supercomplex with light-harvesting complex I | | 分子名称: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Suga, M, Miyazaki, N, Takahashi, Y. | | 登録日 | 2019-03-20 | | 公開日 | 2019-06-19 | | 最終更新日 | 2019-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure of the green algal photosystem I supercomplex with a decameric light-harvesting complex I.
Nat.Plants, 5, 2019
|
|
6GXM
 
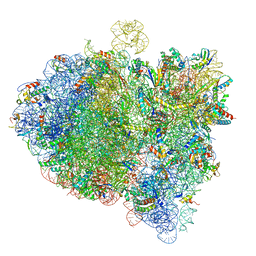 | | Cryo-EM structure of an E. coli 70S ribosome in complex with RF3-GDPCP, RF1(GAQ) and Pint-tRNA (State II) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Graf, M, Huter, P, Maracci, C, Peterek, M, Rodnina, M.V, Wilson, D.N. | | 登録日 | 2018-06-27 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1.
Nat Commun, 9, 2018
|
|
3S0F
 
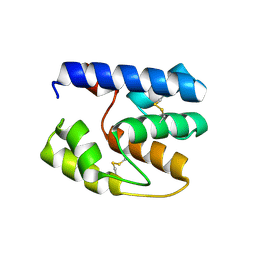 | | Apis mellifera OBP14 native apo, crystal form 2 | | 分子名称: | OBP14 | | 著者 | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | 登録日 | 2011-05-13 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
6GZZ
 
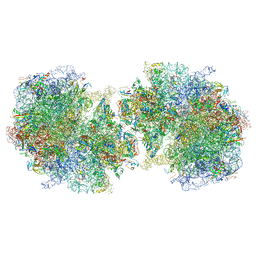 | | T. thermophilus hibernating 100S ribosome (amc) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Flygaard, R.K, Jenner, L.B. | | 登録日 | 2018-07-05 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.13 Å) | | 主引用文献 | Cryo-EM structure of the hibernating Thermus thermophilus 100S ribosome reveals a protein-mediated dimerization mechanism.
Nat Commun, 9, 2018
|
|
6H4N
 
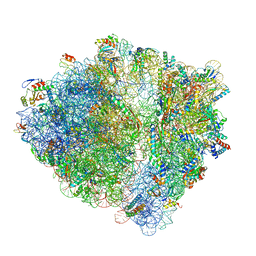 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - 70S Hibernating E. coli Ribosome | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | 著者 | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, N.D. | | 登録日 | 2018-07-22 | | 公開日 | 2018-09-05 | | 最終更新日 | 2018-10-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
6HCM
 
 | | Structure of the rabbit collided di-ribosome (stalled monosome) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | 著者 | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | 登録日 | 2018-08-15 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6.8 Å) | | 主引用文献 | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
3T5H
 
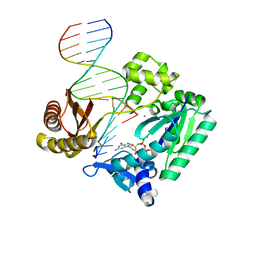 | | Ternary complex of HNE Adduct modified DNA (5'-CXG-3' vs 13-mer) with Dpo4 and incoming dDGT | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*AP*CP*(HN1)P*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | 著者 | Banerjee, S, Christov, P.P, Kozekova, A, Rizzo, C.J, Stone, M.P. | | 登録日 | 2011-07-27 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Replication bypass of the trans-4-Hydroxynonenal-derived (6S,8R,11S)-1,N(2)-deoxyguanosine DNA adduct by the sulfolobus solfataricus DNA polymerase IV.
Chem.Res.Toxicol., 25, 2012
|
|
6JW4
 
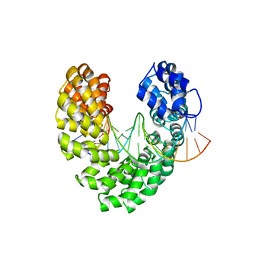 | | Degenerate RVD RG forms a distinct loop conformation | | 分子名称: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5HC)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | 著者 | Liu, L, Yi, C. | | 登録日 | 2019-04-18 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
6JW3
 
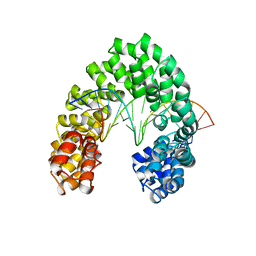 | | Degenerate RVD RG forms a distinct loop conformation | | 分子名称: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5CM)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | 著者 | Liu, L, Yi, C. | | 登録日 | 2019-04-18 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
3QDZ
 
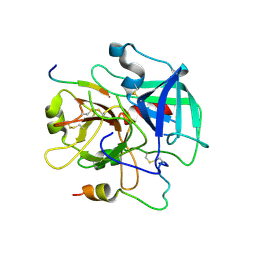 | | Crystal structure of the human thrombin mutant D102N in complex with the extracellular fragment of human PAR4. | | 分子名称: | Proteinase-activated receptor 4, Thrombin heavy chain, Thrombin light chain | | 著者 | Gandhi, P, Chen, Z, Appelbaum, E, Zapata, F, Di Cera, E. | | 登録日 | 2011-01-19 | | 公開日 | 2011-06-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of thrombin-protease-receptor interactions
IUBMB LIFE, 63, 2011
|
|
6HL0
 
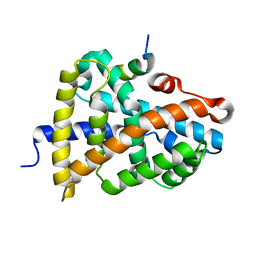 | | Crystal Structure of Farnesoid X receptor (FXR) with bound NCoA-2 peptide | | 分子名称: | Bile acid receptor, NCoA-2 peptide (Nuclear receptor coactivator 2), LYS-GLU-ASN-ALA-LEU-LEU-ARG-TYR-LEU-LEU-ASP-LYS-ASP | | 著者 | Kudlinzki, D, Merk, D, Linhard, V.L, Saxena, K, Schubert-Zsilavecz, M, Schwalbe, H. | | 登録日 | 2018-09-10 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Molecular tuning of farnesoid X receptor partial agonism.
Nat Commun, 10, 2019
|
|
6ICZ
 
 | | Cryo-EM structure of a human post-catalytic spliceosome (P complex) at 3.0 angstrom | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | 著者 | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | 登録日 | 2018-09-07 | | 公開日 | 2019-03-13 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6IP6
 
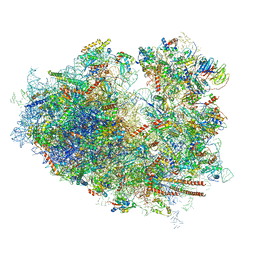 | | Cryo-EM structure of the CMV-stalled human 80S ribosome with HCV IRES (Structure iii) | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | 登録日 | 2018-11-02 | | 公開日 | 2019-05-29 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
6JW2
 
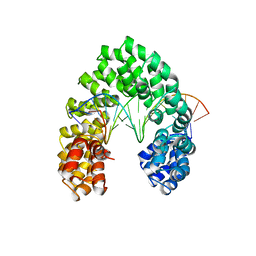 | | Universal RVD R* accommodates 5hmC via water-mediated interactions | | 分子名称: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5HC)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | 著者 | Liu, L, Yi, C. | | 登録日 | 2019-04-18 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.03 Å) | | 主引用文献 | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
3Q91
 
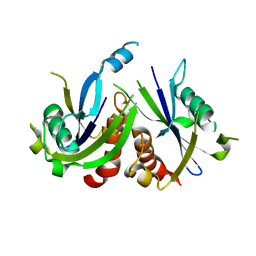 | | Crystal Structure of Human Uridine Diphosphate Glucose Pyrophosphatase (NUDT14) | | 分子名称: | Uridine diphosphate glucose pyrophosphatase | | 著者 | Tresaugues, L, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Thorsell, A.G, Van Der Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | 登録日 | 2011-01-07 | | 公開日 | 2011-02-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structure of Human Uridine Diphosphate Glucose Pyrophosphatase (NUDT14)
To be Published
|
|
6KF3
 
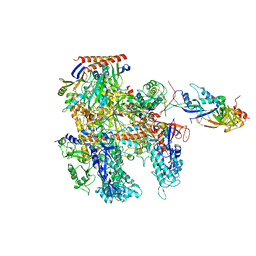 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | 分子名称: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | 著者 | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | 登録日 | 2019-07-06 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
3UBR
 
 | | Laue structure of Shewanella oneidensis cytochrome-c Nitrite Reductase | | 分子名称: | CALCIUM ION, Cytochrome c-552, HEME C | | 著者 | Youngblut, M, Judd, E.T, Srajer, V, Sayed, B, Goeltzner, T, Elliott, S, Schmidt, M, Pacheco, A. | | 登録日 | 2011-10-24 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Laue crystal structure of Shewanella oneidensis cytochrome c nitrite reductase from a high-yield expression system.
J.Biol.Inorg.Chem., 17, 2012
|
|
5OVK
 
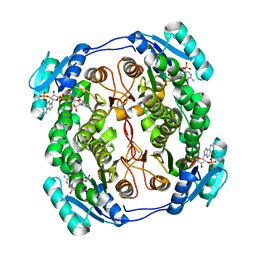 | | Crystal structure MabA bound to NADPH from M. smegmatis | | 分子名称: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Kussau, T, Van Wyk, N, Viljoen, A, Olieric, V, Flipo, M, Kremer, L, Blaise, M. | | 登録日 | 2017-08-29 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural rearrangements occurring upon cofactor binding in the Mycobacterium smegmatis beta-ketoacyl-acyl carrier protein reductase MabA.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
