8XZH
 
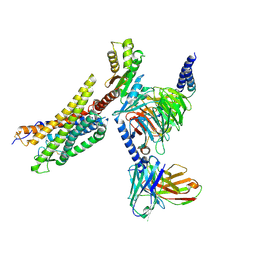 | | Cryo-EM structure of the MM07-bound human APLNR-Gi complex | | 分子名称: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, W, Ji, S, Zhang, Y. | | 登録日 | 2024-01-21 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZI
 
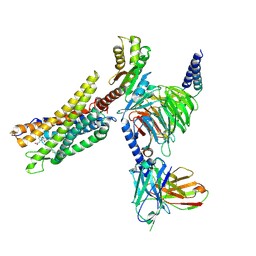 | | Cryo-EM structure of the CMF-019-bound human APLNR-Gi complex | | 分子名称: | (3~{S})-5-methyl-3-[[1-pentan-3-yl-2-(thiophen-2-ylmethyl)benzimidazol-5-yl]carbonylamino]hexanoic acid, Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Wang, W, Ji, S, Zhang, Y. | | 登録日 | 2024-01-21 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
6TTV
 
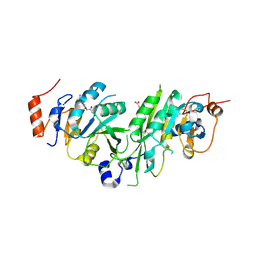 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 3 (ASI_M3M_138) | | 分子名称: | (2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolane-2-carboxamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-12-30 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
3EWF
 
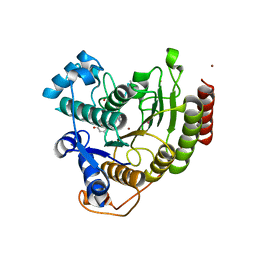 | | Crystal Structure Analysis of human HDAC8 H143A variant complexed with substrate. | | 分子名称: | 7-AMINO-4-METHYL-CHROMEN-2-ONE, Histone deacetylase 8, PEPTIDIC SUBSTRATE, ... | | 著者 | Dowling, D.P, Gantt, S.L, Gattis, S.G, Fierke, C.A, Christianson, D.W. | | 登録日 | 2008-10-14 | | 公開日 | 2008-12-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors.
Biochemistry, 47, 2008
|
|
9AZ9
 
 | | Chloride Sites in Photoactive Yellow Protein (Chloride-Free Reference Structure) | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | 著者 | Dyda, F, Schotte, F, Anfinrud, P, Cho, H.S. | | 登録日 | 2024-03-10 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Watching a signaling protein function: What has been learned over four decades of time-resolved studies of photoactive yellow protein.
Struct Dyn., 11, 2024
|
|
3EWI
 
 | | Structural analysis of the C-terminal domain of murine CMP-Sialic acid Synthetase | | 分子名称: | N-acylneuraminate cytidylyltransferase | | 著者 | Oschlies, M, Dickmanns, A, Stummeyer, K, Gerardy-Schahn, R, Ficner, R, Muenster-Kuehnel, A.K. | | 登録日 | 2008-10-15 | | 公開日 | 2009-08-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A C-terminal phosphatase module conserved in vertebrate CMP-sialic acid synthetases provides a tetramerization interface for the physiologically active enzyme.
J.Mol.Biol., 393, 2009
|
|
8D47
 
 | |
8XZG
 
 | | Cryo-EM structure of the [Pyr1]-apelin-13-bound human APLNR-Gi complex | | 分子名称: | Apelin receptor, Apelin-13, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Wang, W, Ji, S, Zhang, Y. | | 登録日 | 2024-01-21 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
7W93
 
 | | Crystal structure of E.coli pseudouridine kinase PsuK complexed with N1-methyl-pseudouridine | | 分子名称: | 5-[(2S,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1-methyl-pyrimidine-2,4-dione, POTASSIUM ION, PfkB domain protein | | 著者 | Li, K.J, Li, X.J, Wu, B.X. | | 登録日 | 2021-12-09 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure Characterization of Escherichia coli Pseudouridine Kinase PsuK.
Front Microbiol, 13, 2022
|
|
8YJX
 
 | |
6TJH
 
 | | Crystal structure of the computationally designed Cake9 protein | | 分子名称: | Cake9, GLYCEROL, SULFATE ION | | 著者 | Mylemans, B, Laier, I, Noguchi, H, Voet, A.R.D. | | 登録日 | 2019-11-26 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
6NR9
 
 | | hTRiC-hPFD Class5 | | 分子名称: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | 著者 | Gestaut, D, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | 登録日 | 2019-01-23 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (8.5 Å) | | 主引用文献 | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
5MB3
 
 | |
6NRC
 
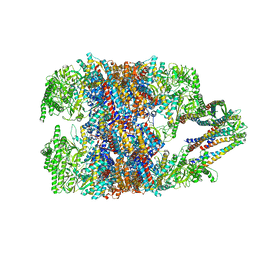 | | hTRiC-hPFD Class3 | | 分子名称: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | 著者 | Gestaut, D, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | 登録日 | 2019-01-23 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (8.3 Å) | | 主引用文献 | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
6TJC
 
 | | Crystal structure of the computationally designed Cake3 protein | | 分子名称: | Cake3, GLYCEROL, PHOSPHATE ION | | 著者 | Laier, I, Mylemans, B, Voet, A.R.D, Noguchi, H. | | 登録日 | 2019-11-26 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
3F7R
 
 | |
6TJG
 
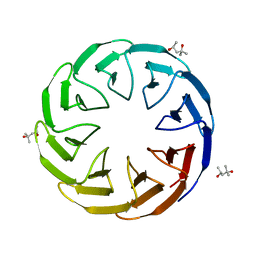 | | Crystal structure of the computationally designed Cake8 protein | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cake8 | | 著者 | Laier, I, Mylemans, B, Noguchi, H, Voet, A.R.D. | | 登録日 | 2019-11-26 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
5LIP
 
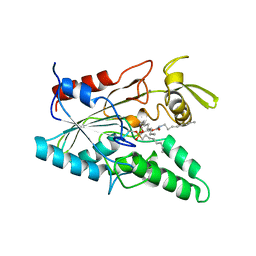 | | PSEUDOMONAS LIPASE COMPLEXED WITH RC-(RP, SP)-1,2-DIOCTYLCARBAMOYLGLYCERO-3-O-OCTYLPHOSPHONATE | | 分子名称: | CALCIUM ION, OCTYL-PHOSPHINIC ACID 1,2-BIS-OCTYLCARBAMOYLOXY-ETHYL ESTER, TRIACYL-GLYCEROL HYDROLASE | | 著者 | Lang, D.A, Dijkstra, B.W. | | 登録日 | 1997-09-02 | | 公開日 | 1998-08-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of the chiral selectivity of Pseudomonas cepacia lipase
Eur.J.Biochem., 254, 1998
|
|
8YD8
 
 | |
8YBX
 
 | |
6NRD
 
 | | hTRiC-hPFD Class4 | | 分子名称: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | 著者 | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | 登録日 | 2019-01-23 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (8.2 Å) | | 主引用文献 | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
5MB6
 
 | |
3FKU
 
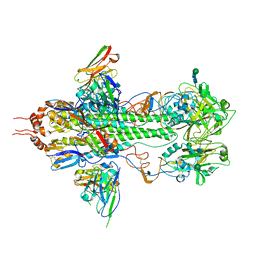 | | Crystal structure of influenza hemagglutinin (H5) in complex with a broadly neutralizing antibody F10 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Neutralizing antibody F10, ... | | 著者 | Hwang, W.C, Santelli, E, Stec, B, Wei, G, Cadwell, G, Bankston, L.A, Sui, J, Perez, S, Aird, D, Chen, L.M, Ali, M, Murakami, A, Yammanuru, A, Han, T, Cox, N, Donis, R.O, Liddington, R.C, Marasco, W.A. | | 登録日 | 2008-12-17 | | 公開日 | 2009-02-24 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses.
Nat.Struct.Mol.Biol., 16, 2009
|
|
5MCQ
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE AND EXOSITE BINDING PEPTIDE INHIBITOR | | 分子名称: | 1,2-ETHANEDIOL, BACE-1 ACTIVE AND EXOSITE BINDING INHIBITOR, Beta-secretase 1 | | 著者 | Kuglstatter, A, Stihle, M, Benz, J. | | 登録日 | 2016-11-10 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
6TBM
 
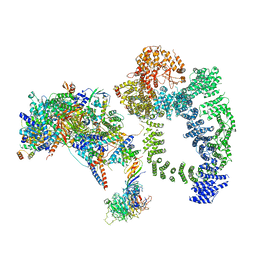 | | Structure of SAGA bound to TBP, including Spt8 and DUB | | 分子名称: | Polyubiquitin-B, SAGA-associated factor 11, Spt20, ... | | 著者 | Papai, G, Frechard, A, Kolesnikova, O, Crucifix, C, Schultz, P, Ben-Shem, A. | | 登録日 | 2019-11-01 | | 公開日 | 2020-02-12 | | 最終更新日 | 2021-06-30 | | 実験手法 | ELECTRON MICROSCOPY (20 Å) | | 主引用文献 | Structure of SAGA and mechanism of TBP deposition on gene promoters.
Nature, 577, 2020
|
|
