4ARZ
 
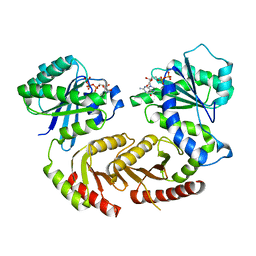 | | The crystal structure of Gtr1p-Gtr2p complexed with GTP-GDP | | 分子名称: | GTP-BINDING PROTEIN GTR1, GTP-BINDING PROTEIN GTR2, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Jeong, J.H, Kim, Y.G. | | 登録日 | 2012-04-27 | | 公開日 | 2012-07-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal Structure of the Gtr1Pgtp-Gtr2Pgdp Complex Reveals Large Structural Rearrangements Triggered by GTP-to-Gdp Conversion
J.Biol.Chem., 287, 2012
|
|
7ZMU
 
 | |
4WSZ
 
 | |
4WT0
 
 | |
7AOG
 
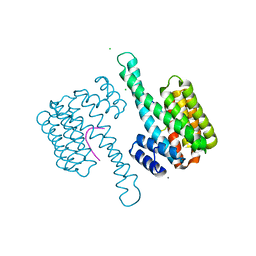 | | 14-3-3 sigma in complex with Pin1 binding site pS72 | | 分子名称: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | 登録日 | 2020-10-14 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
6VCD
 
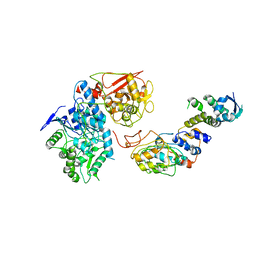 | | Cryo-EM structure of IRP2-FBXL5-SKP1 complex | | 分子名称: | F-box/LRR-repeat protein 5, FE2/S2 (INORGANIC) CLUSTER, Iron-responsive element binding protein 2, ... | | 著者 | Wang, H, Shi, H, Zheng, N. | | 登録日 | 2019-12-20 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | FBXL5 Regulates IRP2 Stability in Iron Homeostasis via an Oxygen-Responsive [2Fe2S] Cluster.
Mol.Cell, 78, 2020
|
|
6POR
 
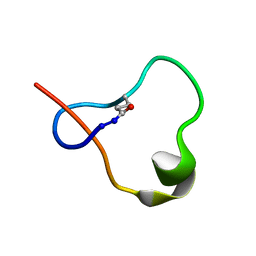 | |
8K6Z
 
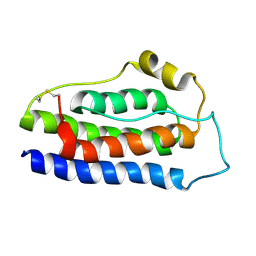 | | NMR structure of human leptin | | 分子名称: | Leptin | | 著者 | Fan, X, Qin, R, Yuan, W, Fan, J, Huang, W, Lin, Z. | | 登録日 | 2023-07-26 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of human leptin reveals a conformational plasticity important for receptor recognition.
Structure, 32, 2024
|
|
7BG3
 
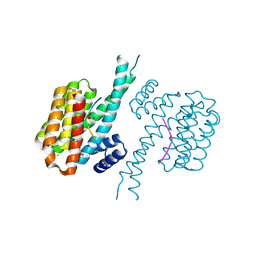 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound PC2046 | | 分子名称: | 1-(3-bromanyl-4-methyl-phenyl)-2-(2-bromophenyl)imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | 著者 | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | 登録日 | 2021-01-05 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
5XQM
 
 | |
1C20
 
 | |
8FP5
 
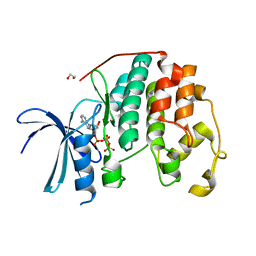 | | CDK2 liganded with ATP and Mg2+ | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cyclin-dependent kinase 2, ... | | 著者 | Schonbrunn, E, Sun, L. | | 登録日 | 2023-01-04 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
1B4T
 
 | | H48C YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | 分子名称: | CHLORIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | 著者 | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | 登録日 | 1998-12-23 | | 公開日 | 1999-12-23 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1B4L
 
 | | 15 ATMOSPHERE OXYGEN YEAST CU/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | 分子名称: | COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ZINC ION | | 著者 | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | 登録日 | 1998-12-22 | | 公開日 | 1999-12-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1YFD
 
 | | Crystal structure of the Y122H mutant of ribonucleotide reductase R2 protein from E. coli | | 分子名称: | MERCURY (II) ION, MU-OXO-DIIRON, Ribonucleoside-diphosphate reductase 1 beta chain | | 著者 | Kolberg, M, Logan, D.T, Bleifuss, G, Poetsch, S, Sjoeberg, B.M, Graeslund, A, Lubitz, W, Lassmann, G, Lendzian, F. | | 登録日 | 2004-12-31 | | 公開日 | 2005-02-15 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A new tyrosyl radical on Phe208 as ligand to the diiron center in Escherichia coli ribonucleotide reductase, mutant R2-Y122H. Combined x-ray diffraction and EPR/ENDOR studies
J.Biol.Chem., 280, 2005
|
|
7RWF
 
 | | Crystal structure of CDK2 in complex with TW8672 | | 分子名称: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, Cyclin-dependent kinase 2 | | 著者 | Sun, L, Schonbrunn, E. | | 登録日 | 2021-08-19 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
7S84
 
 | | Crystal structure of CDK2 liganded with compound TW8972 | | 分子名称: | 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | 著者 | Sun, L, Schonbrunn, E. | | 登録日 | 2021-09-17 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
4MHG
 
 | | Crystal structure of ETV6 bound to a specific DNA sequence | | 分子名称: | Complementary Specific 14 bp DNA, Specific 14 bp DNA, Transcription factor ETV6 | | 著者 | Chan, A.C, De, S, Coyne III, H.J, Okon, M, Murphy, M.E, Graves, B.J, McIntosh, L.P. | | 登録日 | 2013-08-29 | | 公開日 | 2014-01-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | Steric Mechanism of Auto-Inhibitory Regulation of Specific and Non-Specific DNA Binding by the ETS Transcriptional Repressor ETV6.
J.Mol.Biol., 426, 2014
|
|
2LSV
 
 | | The NMR high resolution structure of yeast Tah1 in complex with the Hsp90 C-terminal tail | | 分子名称: | ATP-dependent molecular chaperone HSP82, TPR repeat-containing protein associated with Hsp90 | | 著者 | Back, R, Dominguez, C, Rothe, B, Bobo, C, Beaufils, C, Morera, S, Meyer, P, Charpentier, B, Branlant, C, Allain, F, Manival, X. | | 登録日 | 2012-05-07 | | 公開日 | 2013-05-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | High-Resolution Structural Analysis Shows How Tah1 Tethers Hsp90 to the R2TP Complex.
Structure, 21, 2013
|
|
6ZV1
 
 | |
6ZV2
 
 | | TFIIS N-terminal domain (TND) from human PPP1R10 | | 分子名称: | Serine/threonine-protein phosphatase 1 regulatory subunit 10 | | 著者 | Veverka, V. | | 登録日 | 2020-07-23 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A ubiquitous disordered protein interaction module orchestrates transcription elongation.
Science, 374, 2021
|
|
6ZV4
 
 | |
6ZV0
 
 | |
6E8E
 
 | |
3QWR
 
 | |
