5I73
 
 | | X-ray structure of the ts3 human serotonin transporter complexed with s-citalopram at the central and allosteric sites | | 分子名称: | (1S)-1-[3-(dimethylamino)propyl]-1-(4-fluorophenyl)-1,3-dihydro-2-benzofuran-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-AMINOHEXANOIC ACID, ... | | 著者 | Coleman, J.A, Green, E.M, Gouaux, E. | | 登録日 | 2016-02-16 | | 公開日 | 2016-04-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.24 Å) | | 主引用文献 | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
4X4A
 
 | | Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with 2,7-Anhydro-Neu5Ac | | 分子名称: | 2-ACETYLAMINO-7-(1,2-DIHYDROXY-ETHYL)-3-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCTANE-5-CARBOXYLIC ACID, ACETYL GROUP, Anhydrosialidase, ... | | 著者 | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | 登録日 | 2014-12-02 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation.
Nat Commun, 6, 2015
|
|
6OBL
 
 | | JAK2 JH2 in complex with JAK168 | | 分子名称: | Tyrosine-protein kinase JAK2, [4-({5-amino-3-[(4-cyanophenyl)amino]-1H-1,2,4-triazole-1-carbonyl}amino)phenoxy]acetic acid | | 著者 | Krimmer, S.G, Liosi, M.E, Schlessinger, J, Jorgensen, W.L. | | 登録日 | 2019-03-21 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.061 Å) | | 主引用文献 | Selective Janus Kinase 2 (JAK2) Pseudokinase Ligands with a Diaminotriazole Core.
J.Med.Chem., 63, 2020
|
|
6R7Y
 
 | | CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (low Ca2+, closed form) | | 分子名称: | Anoctamin-10, CALCIUM ION | | 著者 | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | 登録日 | 2019-03-29 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
8OGX
 
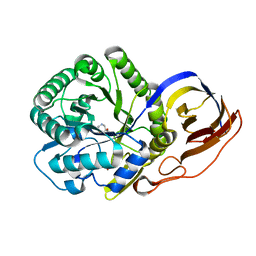 | | Beta-glucuronidase from Acidobacterium capsulatum in complex with inhibitor R3794 | | 分子名称: | (3~{S},4~{S})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, PHOSPHATE ION, beta-glucuronidase from Acidobacterium capsulatum | | 著者 | Moran, E.M, Davies, G.J, Chen, C, Nieuwendijk, E.V, Wu, L, Skoulikopoulou, F, Riet, V.V, Overkleeft, H.S, Armstrong, Z. | | 登録日 | 2023-03-20 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
1S9G
 
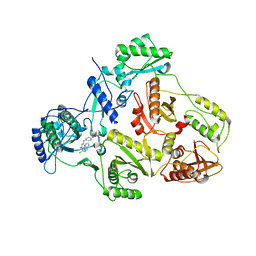 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R120394. | | 分子名称: | 4-[4-AMINO-6-(5-CHLORO-1H-INDOL-4-YLMETHYL)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | 著者 | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | 登録日 | 2004-02-04 | | 公開日 | 2004-05-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
7NYD
 
 | | cryoEM structure of 2C9-sMAC | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Menny, A, Couves, E.C, Bubeck, D. | | 登録日 | 2021-03-22 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Structural basis of soluble membrane attack complex packaging for clearance.
Nat Commun, 12, 2021
|
|
1M3V
 
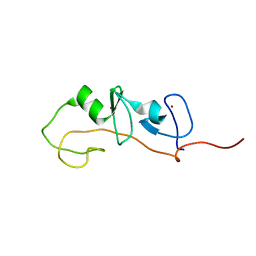 | | FLIN4: Fusion of the LIM binding domain of Ldb1 and the N-terminal LIM domain of LMO4 | | 分子名称: | ZINC ION, fusion of the LIM interacting domain of ldb1 and the N-terminal LIM domain of LMO4 | | 著者 | Deane, J.E, Mackay, J.P, Kwan, A.H.Y, Sum, E.Y, Visvader, J.E, Matthews, J.M. | | 登録日 | 2002-06-30 | | 公開日 | 2003-05-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4
EMBO J., 22, 2003
|
|
5N0Z
 
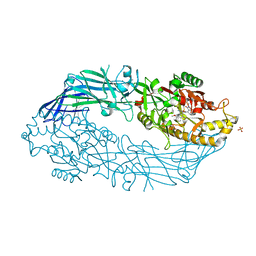 | | hPAD4 crystal complex with AFM-41a | | 分子名称: | 2-ethyl-~{N}-[(1~{S})-4-(2-fluoranylethanimidoylamino)-1-(4-methoxy-1-methyl-benzimidazol-2-yl)butyl]-3-oxidanylidene-1~{H}-isoindole-4-carboxamide, CALCIUM ION, Protein-arginine deiminase type-4, ... | | 著者 | Beaumont, E, Kerry, P, Thompson, P, Muth, A, Subramanian, V, Nagar, M, Srinath, H, Clancy, K, Parelkar, S. | | 登録日 | 2017-02-03 | | 公開日 | 2017-05-24 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Development of a Selective Inhibitor of Protein Arginine Deiminase 2.
J. Med. Chem., 60, 2017
|
|
6O6C
 
 | | RNA polymerase II elongation complex arrested at a CPD lesion | | 分子名称: | DNA (27-MER), DNA (5'-D(P*GP*GP*AP*GP*AP*AP*GP*GP*AP*GP*CP*AP*GP*AP*GP*C)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Lahiri, I, Leshziner, A.E. | | 登録日 | 2019-03-05 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | 3.1 angstrom structure of yeast RNA polymerase II elongation complex stalled at a cyclobutane pyrimidine dimer lesion solved using streptavidin affinity grids.
J.Struct.Biol., 207, 2019
|
|
5N1B
 
 | | hPAD4 crystal complex with AFM-14a | | 分子名称: | CALCIUM ION, Protein-arginine deiminase type-4, SULFATE ION, ... | | 著者 | Beaumont, E, Kerry, P, Thompson, P, Muth, A, Subramanian, V, Nagar, M, Srinath, H, Clancy, K, Parelkar, S. | | 登録日 | 2017-02-06 | | 公開日 | 2017-05-24 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Development of a Selective Inhibitor of Protein Arginine Deiminase 2.
J. Med. Chem., 60, 2017
|
|
6R60
 
 | |
8OI4
 
 | | Metagenomic Beta-galactosidase from Glycoside Hydrolase family GH154 | | 分子名称: | Beta-galactosidase, CHLORIDE ION, GLYCEROL | | 著者 | Pijning, T, Hameleers, L, Jurak, E, Guskov, A. | | 登録日 | 2023-03-22 | | 公開日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Novel beta-galactosidase activity and first crystal structure of Glycoside Hydrolase family 154.
N Biotechnol, 80, 2023
|
|
6FPW
 
 | | Structure of fully reduced Hydrogenase (Hyd-1) | | 分子名称: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, FE3-S4 CLUSTER, ... | | 著者 | Carr, S.B, Armstrong, F.A, Evans, R.M. | | 登録日 | 2018-02-12 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6G95
 
 | | Crystal structure of Ebolavirus glycoprotein in complex with thioridazine | | 分子名称: | 10-{2-[(2R)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Zhao, Y, Ren, J, Fry, E.E, Xiao, J, Townsend, A.R, Stuart, D.I. | | 登録日 | 2018-04-10 | | 公開日 | 2018-05-23 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structures of Ebola Virus Glycoprotein Complexes with Tricyclic Antidepressant and Antipsychotic Drugs.
J. Med. Chem., 61, 2018
|
|
7UKV
 
 | | Wild type EGFR in complex with Lazertinib (YH25448) | | 分子名称: | Epidermal growth factor receptor, N-[5-{[(4P)-4-{4-[(dimethylamino)methyl]-3-phenyl-1H-pyrazol-1-yl}pyrimidin-2-yl]amino}-4-methoxy-2-(morpholin-4-yl)phenyl]propanamide | | 著者 | Beyett, T.S, Pham, C, Eck, M.J, Heppner, D.E. | | 登録日 | 2022-04-02 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis for Inhibition of Mutant EGFR with Lazertinib (YH25448).
Acs Med.Chem.Lett., 13, 2022
|
|
8OGD
 
 | | Structure of zinc(II) double mutant human carbonic anhydrase II bound to thiocyanate | | 分子名称: | 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, THIOCYANATE ION, ... | | 著者 | Silva, J.M, Cerofolini, L, Carvalho, A.L, Ravera, E, Fragai, M, Parigi, G, Macedo, A.L, Geraldes, C.F.G.C, Luchinat, C. | | 登録日 | 2023-03-20 | | 公開日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Elucidating the concentration-dependent effects of thiocyanate binding to carbonic anhydrase.
J.Inorg.Biochem., 244, 2023
|
|
6LYF
 
 | |
8OGE
 
 | | Structure of cobalt(II) substituted double mutant human carbonic anhydrase II bound to thiocyanate | | 分子名称: | 4-(HYDROXYMERCURY)BENZOIC ACID, COBALT (II) ION, Carbonic anhydrase 2, ... | | 著者 | Silva, J.M, Cerofolini, L, Carvalho, A.L, Ravera, E, Fragai, M, Parigi, G, Macedo, A.L, Geraldes, C.F.G.C, Luchinat, C. | | 登録日 | 2023-03-20 | | 公開日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Elucidating the concentration-dependent effects of thiocyanate binding to carbonic anhydrase.
J.Inorg.Biochem., 244, 2023
|
|
5ECY
 
 | |
3I2B
 
 | | The crystal structure of human 6 Pyruvoyl Tetrahydrobiopterin Synthase | | 分子名称: | 1,2-ETHANEDIOL, 6-pyruvoyl tetrahydrobiopterin synthase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Ugochukwu, E, Cocking, R, Pilka, E, Yue, W.W, Bray, J.E, Chaikuad, A, Krojer, T, Muniz, J, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | 登録日 | 2009-06-29 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The crystal structure of human 6 Pyruvoyl Tetrahydrobiopterin Synthase
To be Published
|
|
2GHP
 
 | | Crystal structure of the N-terminal 3 RNA binding domains of the yeast splicing factor Prp24 | | 分子名称: | U4/U6 snRNA-associated splicing factor PRP24 | | 著者 | Bae, E, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2006-03-27 | | 公開日 | 2006-04-25 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure and interactions of the first three RNA recognition motifs of splicing factor prp24.
J.Mol.Biol., 367, 2007
|
|
5EEB
 
 | | Apo form of thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 | | 分子名称: | Aldehyde dehydrogenase | | 著者 | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | 登録日 | 2015-10-22 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.038 Å) | | 主引用文献 | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
6Q9Y
 
 | |
6PZ0
 
 | | Crystal structure of oxidized iodotyrosine deiodinase (IYD) bound to FMN and L-Tyrosine | | 分子名称: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, TYROSINE, ... | | 著者 | Sun, Z, Kavran, J.M, Rokita, S.E. | | 登録日 | 2019-07-31 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The minimal structure for iodotyrosine deiodinase function is defined by an outlier protein from the thermophilic bacterium Thermotoga neapolitana.
J.Biol.Chem., 297, 2021
|
|
