2R2T
 
 | | d(ATTTAGTTAACTAAAT) complexed with MMLV RT catalytic fragment | | 分子名称: | DNA (5'-D(*DAP*DTP*DTP*DTP*DAP*DGP*DTP*DT)-3'), DNA (5'-D(P*DAP*DAP*DCP*DTP*DAP*DAP*DAP*DT)-3'), Reverse transcriptase | | 著者 | Goodwin, K.D, Lewis, M.A, Long, E.C, Georgiadis, M.M. | | 登録日 | 2007-08-27 | | 公開日 | 2008-07-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of DNA-bound Co(III) bleomycin B2: Insights on intercalation and minor groove binding.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7L7J
 
 | | Cryo-EM structure of Hsp90:p23 closed-state complex | | 分子名称: | Heat shock protein HSP 90-alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Prostaglandin E synthase 3 | | 著者 | Lee, K, Thwin, A.C, Tse, E, Gates, S.N, Southworth, D.R. | | 登録日 | 2020-12-28 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state.
Mol.Cell, 81, 2021
|
|
7L7I
 
 | | Cryo-EM structure of Hsp90:FKBP51:p23 closed-state complex | | 分子名称: | Heat shock protein HSP 90-alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | 著者 | Lee, K, Thwin, A.C, Tse, E, Gates, S.N, Southworth, D.R. | | 登録日 | 2020-12-28 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state.
Mol.Cell, 81, 2021
|
|
2R2S
 
 | | Co(III)bleomycinB2 bound to d(ATTAGTTATAACTAAT) complexed with MMLV RT catalytic fragment | | 分子名称: | BLEOMYCIN B2, COBALT (III) ION, DNA (5'-D(*DAP*DTP*DTP*DAP*DGP*DTP*DT)-3'), ... | | 著者 | Goodwin, K.D, Lewis, M.A, Long, E.C, Georgiadis, M.M. | | 登録日 | 2007-08-27 | | 公開日 | 2008-07-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of DNA-bound Co(III) bleomycin B2: Insights on intercalation and minor groove binding.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5EAX
 
 | | Crystal structure of Dna2 in complex with an ssDNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA replication ATP-dependent helicase/nuclease DNA2, ... | | 著者 | Zhou, C, Pourmal, S, Pavletich, N.P. | | 登録日 | 2015-10-17 | | 公開日 | 2015-11-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Dna2 nuclease-helicase structure, mechanism and regulation by Rpa.
Elife, 4, 2015
|
|
5EAN
 
 | | Crystal structure of Dna2 in complex with a 5' overhang DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*AP*CP*TP*CP*TP*GP*CP*CP*AP*AP*GP*AP*GP*GP*A)-3'), ... | | 著者 | Zhou, C, Pourmal, S, Pavletich, N.P. | | 登録日 | 2015-10-16 | | 公開日 | 2015-11-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Dna2 nuclease-helicase structure, mechanism and regulation by Rpa.
Elife, 4, 2015
|
|
1C1B
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GCA-186 | | 分子名称: | 6-(3',5'-DIMETHYLBENZYL)-1-ETHOXYMETHYL-5-ISOPROPYLURACIL, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | 著者 | Hopkins, A.L, Ren, J, Tanaka, H, Baba, B, Okamato, M, Stuart, D.I, Stammers, D.K. | | 登録日 | 1999-07-21 | | 公開日 | 2000-07-21 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Design of MKC-442 (emivirine) analogues with improved activity against drug-resistant HIV mutants.
J.Med.Chem., 42, 1999
|
|
1DTT
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 (PETT130A94) | | 分子名称: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | 著者 | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | 登録日 | 2000-01-13 | | 公開日 | 2000-04-02 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1DTQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-1 (PETT131A94) | | 分子名称: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-NITRILOMETHYL-PYRIDYL]-THIOUREA | | 著者 | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | 登録日 | 2000-01-13 | | 公開日 | 2000-03-20 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
8DAW
 
 | |
8DAU
 
 | |
8DAR
 
 | |
8DAS
 
 | |
8DAT
 
 | |
8DAV
 
 | |
5EAW
 
 | | Crystal structure of Dna2 nuclease-helicase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA replication ATP-dependent helicase/nuclease DNA2, IRON/SULFUR CLUSTER | | 著者 | Zhou, C, Pourmal, S, Pavletich, N.P. | | 登録日 | 2015-10-17 | | 公開日 | 2015-11-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Dna2 nuclease-helicase structure, mechanism and regulation by Rpa.
Elife, 4, 2015
|
|
1C0T
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH BM+21.1326 | | 分子名称: | (R)-(+)9B-(3-METHYL)PHENYL-2,3-DIHYDROTHIAZOLO[2,3-A]ISOINDOL-5(9BH)-ONE, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | 著者 | Ren, J, Esnouf, R.M, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | 登録日 | 1999-07-19 | | 公開日 | 2000-07-19 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystallographic analysis of the binding modes of thiazoloisoindolinone non-nucleoside inhibitors to HIV-1 reverse transcriptase and comparison with modeling studies.
J.Med.Chem., 42, 1999
|
|
1RTD
 
 | | STRUCTURE OF A CATALYTIC COMPLEX OF HIV-1 REVERSE TRANSCRIPTASE: IMPLICATIONS FOR NUCLEOSIDE ANALOG DRUG RESISTANCE | | 分子名称: | DNA PRIMER FOR REVERSE TRANSCRIPTASE, DNA TEMPLATE FOR REVERSE TRANSCRIPTASE, MAGNESIUM ION, ... | | 著者 | Chopra, R, Huang, H, Verdine, G.L, Harrison, S.C. | | 登録日 | 1998-08-26 | | 公開日 | 1998-12-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance.
Science, 282, 1998
|
|
1EP4
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with S-1153 | | 分子名称: | 5-(3,5-DICHLOROPHENYL)THIO-4-ISOPROPYL-1-(PYRIDIN-4-YL-METHYL)-1H-IMIDAZOL-2-YL-METHYL CARBAMATE, HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Ren, J, Nichols, C, Bird, L.E, Fujiwara, T, Suginoto, H, Stuart, D.I, Stammers, D.K. | | 登録日 | 2000-03-27 | | 公開日 | 2000-09-27 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Binding of the second generation non-nucleoside inhibitor S-1153 to HIV-1 reverse transcriptase involves extensive main chain hydrogen bonding.
J.Biol.Chem., 275, 2000
|
|
1EET
 
 | | HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH THE INHIBITOR MSC204 | | 分子名称: | 1-(5-BROMO-PYRIDIN-2-YL)-3-[2-(6-FLUORO-2-HYDROXY-3-PROPIONYL-PHENYL)-CYCLOPROPYL]-UREA, HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Hogberg, M, Sahlberg, C, Engelhardt, P, Noreen, R, Kangasmetsa, J, Johansson, N.G, Oberg, B, Vrang, L, Zhang, H, Sahlberg, B.L, Unge, T, Lovgren, S, Fridborg, K, Backbro, K. | | 登録日 | 2000-02-03 | | 公開日 | 2001-02-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Urea-PETT compounds as a new class of HIV-1 reverse transcriptase inhibitors. 3. Synthesis and further structure-activity relationship studies of PETT analogues.
J.Med.Chem., 42, 1999
|
|
1C1C
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH TNK-6123 | | 分子名称: | 6-(cyclohexylsulfanyl)-1-(ethoxymethyl)-5-(1-methylethyl)pyrimidine-2,4(1H,3H)-dione, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | 著者 | Hopkins, A.L, Ren, J, Tanaka, H, Baba, M, Okamato, M, Stuart, D.I, Stammers, D.K. | | 登録日 | 1999-07-21 | | 公開日 | 2000-07-21 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Design of MKC-442 (emivirine) analogues with improved activity against drug-resistant HIV mutants.
J.Med.Chem., 42, 1999
|
|
1FKO
 
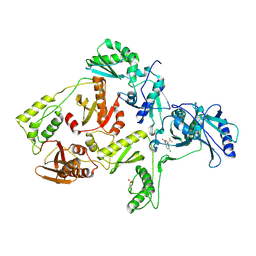 | | CRYSTAL STRUCTURE OF NNRTI RESISTANT K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | 分子名称: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | 著者 | Ren, J, Milton, J, Weaver, K.L, Short, S.A, Stuart, D.I, Stammers, D.K. | | 登録日 | 2000-08-10 | | 公開日 | 2000-11-03 | | 最終更新日 | 2018-03-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase.
Structure Fold.Des., 8, 2000
|
|
1C0U
 
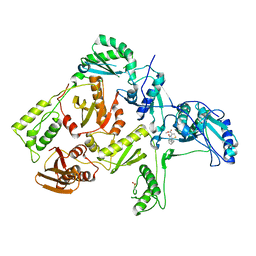 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH BM+50.0934 | | 分子名称: | (R)-(+) 5(9BH)-OXO-9B-PHENYL-2,3-DIHYDROTHIAZOLO[2,3-A]ISOINDOL-3-CARBOXYLIC ACID METHYL ESTER, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | 著者 | Ren, J, Esnouf, R.M, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | 登録日 | 1999-07-19 | | 公開日 | 2000-07-19 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | Crystallographic analysis of the binding modes of thiazoloisoindolinone non-nucleoside inhibitors to HIV-1 reverse transcriptase and comparison with modeling studies.
J.Med.Chem., 42, 1999
|
|
1FK9
 
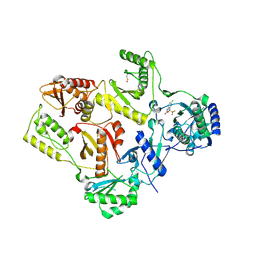 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | 分子名称: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | 著者 | Ren, J, Milton, J, Weaver, K.L, Short, S.A, Stuart, D.I, Stammers, D.K. | | 登録日 | 2000-08-09 | | 公開日 | 2000-11-03 | | 最終更新日 | 2017-02-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase.
STRUCTURE FOLD.DES., 8, 2000
|
|
1FKP
 
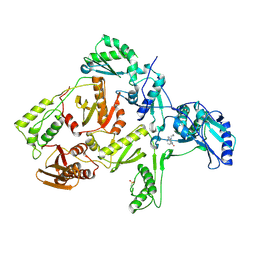 | | CRYSTAL STRUCTURE OF NNRTI RESISTANT K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT, A-CHAIN, ... | | 著者 | Ren, J, Milton, J, Weaver, K.L, Short, S.A, Stuart, D.I, Stammers, D.K. | | 登録日 | 2000-08-10 | | 公開日 | 2000-11-03 | | 最終更新日 | 2018-03-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase.
Structure Fold.Des., 8, 2000
|
|
