7ZAG
 
 | | Cryo-EM structure of a Pyrococcus abyssi 30S bound to Met-initiator tRNA,mRNA, aIF1A and the C-terminal domain of aIF5B. | | 分子名称: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | 著者 | Coureux, P.D, Bourgeois, G, Mechulam, Y, Schmitt, E, Kazan, R. | | 登録日 | 2022-03-22 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
3HFA
 
 | |
3N75
 
 | | X-ray Crystal Structure of the Escherichia coli Inducible Lysine Decarboxylase LdcI | | 分子名称: | GLYCEROL, GUANOSINE-5',3'-TETRAPHOSPHATE, HEXAETHYLENE GLYCOL, ... | | 著者 | Kanjee, U, Alexopoulos, E, Pai, E.F, Houry, W.A. | | 登録日 | 2010-05-26 | | 公開日 | 2011-02-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase.
Embo J., 30, 2011
|
|
7ZAH
 
 | | Cryo-EM structure of a Pyrococcus abyssi 30S bound to Met-initiator tRNA, mRNA, aIF1A and aIF5B | | 分子名称: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | 著者 | Coureux, P.D, Bourgeois, G, Mechulam, Y, Schmitt, E, Kazan, R. | | 登録日 | 2022-03-22 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
7YOZ
 
 | | Cryo-EM structure of human subnucleosome (intermediate form) | | 分子名称: | Histone H3.1, Histone H4, Widom601 DNA FW (145-MER), ... | | 著者 | Nozawa, K, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2022-08-02 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Cryo-electron microscopy structure of the H3-H4 octasome: A nucleosome-like particle without histones H2A and H2B.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YQK
 
 | | cryo-EM structure of gammaH2AXK15ub-H4K20me2 nucleosome bound to 53BP1 | | 分子名称: | DNA (145-MER), Histone H2AX, Histone H2B, ... | | 著者 | Ai, H.S, GuoChao, C, Qingyue, G, Ze-Bin, T, Zhiheng, D, Xin, L, Fan, Y, Ziyu, X, Jia-Bin, L, Changlin, T, Liu, L. | | 登録日 | 2022-08-07 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | Chemical Synthesis of Post-Translationally Modified H2AX Reveals Redundancy in Interplay between Histone Phosphorylation, Ubiquitination, and Methylation on the Binding of 53BP1 with Nucleosomes.
J.Am.Chem.Soc., 144, 2022
|
|
3NBZ
 
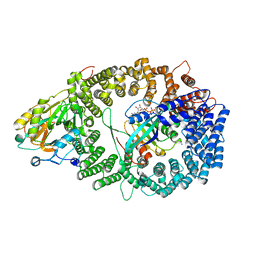 | | Crystal structure of the HIV-1 Rev NES-CRM1-RanGTP nuclear export complex (crystal I) | | 分子名称: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | 登録日 | 2010-06-04 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4Y22
 
 | |
3IE5
 
 | | Crystal structure of Hyp-1 protein from Hypericum perforatum (St John's wort) involved in hypericin biosynthesis | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Michalska, K, Fernandes, H, Sikorski, M.M, Jaskolski, M. | | 登録日 | 2009-07-22 | | 公開日 | 2009-11-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.688 Å) | | 主引用文献 | Crystal structure of Hyp-1, a St. John's wort protein implicated in the biosynthesis of hypericin
J.Struct.Biol., 169, 2010
|
|
3F5P
 
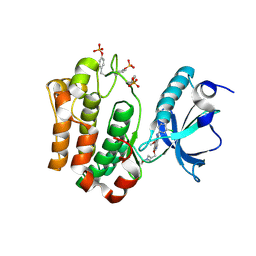 | | Complex Structure of Insulin-like Growth Factor Receptor and 3-Cyanoquinoline Inhibitor | | 分子名称: | 4-[[3-chloro-4-(1-methylimidazol-2-yl)sulfanyl-phenyl]amino]-7-[3-(2-hydroxyethyl-methyl-amino)propoxy]-6-methoxy-quinoline-3-carbonitrile, Insulin-like growth factor 1 receptor | | 著者 | Xu, W, Miller, L.M, Mayer, S.C, Berger, D.M, Boschelli, D.H, Boschelli, F. | | 登録日 | 2008-11-04 | | 公開日 | 2008-12-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Lead identification to generate 3-cyanoquinoline inhibitors of insulin-like growth factor receptor (IGF-1R) for potential use in cancer treatment
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1NLM
 
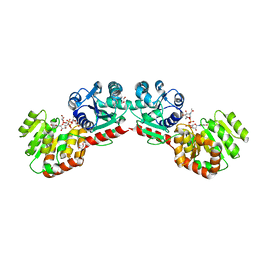 | | CRYSTAL STRUCTURE OF MURG:GLCNAC COMPLEX | | 分子名称: | GLYCEROL, UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | 著者 | Hu, Y, Chen, L, Ha, S, Gross, B, Falcone, B, Walker, D, Mokhtarzadeh, M, Walker, S. | | 登録日 | 2003-01-07 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3NBB
 
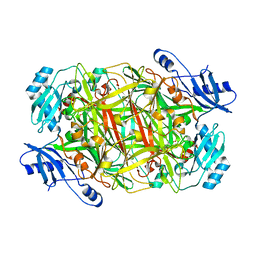 | | Crystal structure of mutant Y305F expressed in E. coli in the copper amine oxidase from hansenula polymorpha | | 分子名称: | COPPER (II) ION, Peroxisomal primary amine oxidase | | 著者 | Chen, Z, Datta, S, DuBois, J.L, Klinman, J.P, Mathews, F.S. | | 登録日 | 2010-06-03 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Mutation at a strictly conserved, active site tyrosine in the copper amine oxidase leads to uncontrolled oxygenase activity.
Biochemistry, 49, 2010
|
|
1OMN
 
 | |
1Q5R
 
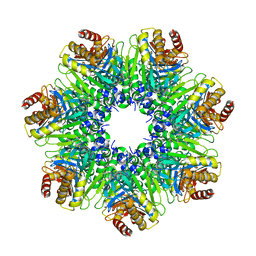 | | The Rhodococcus 20S proteasome with unprocessed pro-peptides | | 分子名称: | proteasome alpha-type subunit 1, proteasome beta-type subunit 1 | | 著者 | Kwon, Y.D, Nagy, I, Adams, P.D, Baumeister, W, Jap, B.K. | | 登録日 | 2003-08-08 | | 公開日 | 2003-12-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly.
J.Mol.Biol., 335, 2004
|
|
3FMW
 
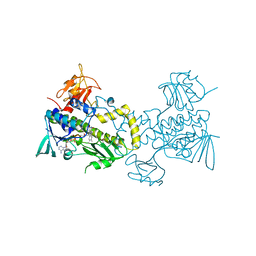 | | The crystal structure of MtmOIV, a Baeyer-Villiger monooxygenase from the mithramycin biosynthetic pathway in Streptomyces argillaceus. | | 分子名称: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Oxygenase | | 著者 | Noinaj, N, Beam, M.P, Wang, C, Rohr, J. | | 登録日 | 2008-12-22 | | 公開日 | 2009-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Crystal structure of Baeyer-Villiger monooxygenase MtmOIV, the key enzyme of the mithramycin biosynthetic pathway .
Biochemistry, 48, 2009
|
|
1P4Z
 
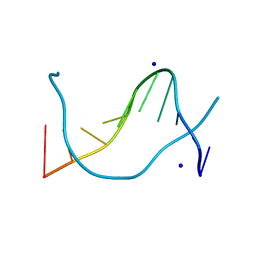 | |
3J3Y
 
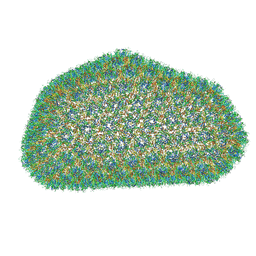 | |
3J9W
 
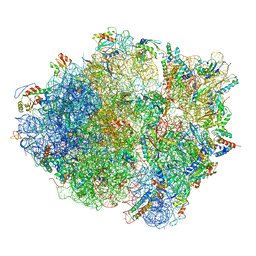 | | Cryo-EM structure of the Bacillus subtilis MifM-stalled ribosome complex | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein bS16, ... | | 著者 | Sohmen, D, Chiba, S, Shimokawa-Chiba, N, Innis, C.A, Berninghausen, O, Beckmann, R, Ito, K, Wilson, D.N. | | 登録日 | 2015-03-16 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling.
Nat Commun, 6, 2015
|
|
6N7P
 
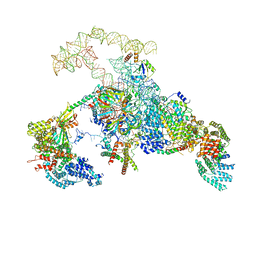 | | S. cerevisiae spliceosomal E complex (UBC4) | | 分子名称: | 56 kDa U1 small nuclear ribonucleoprotein component, Nuclear cap-binding protein complex subunit 1, Nuclear cap-binding protein subunit 2, ... | | 著者 | Liu, S, Li, X, Zhou, Z.H, Zhao, R. | | 登録日 | 2018-11-27 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | A unified mechanism for intron and exon definition and back-splicing.
Nature, 573, 2019
|
|
6N7X
 
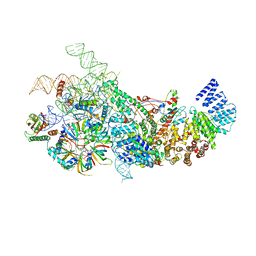 | | S. cerevisiae U1 snRNP | | 分子名称: | 56 kDa U1 small nuclear ribonucleoprotein component, Pre-mRNA-processing factor 39, Protein NAM8, ... | | 著者 | Li, X, Liu, S, Jiang, J, Zhang, L, Espinosa, S, Hill, R.C, Hansen, K.C, Zhou, Z.H, Zhao, R. | | 登録日 | 2018-11-28 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | CryoEM structure of Saccharomyces cerevisiae U1 snRNP offers insight into alternative splicing.
Nat Commun, 8, 2017
|
|
6MTD
 
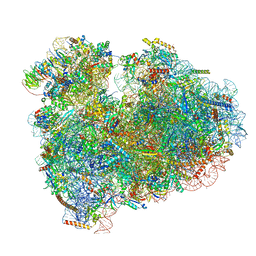 | | Rabbit 80S ribosome with eEF2 and SERBP1 (unrotated state with 40S head swivel) | | 分子名称: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | 著者 | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | 登録日 | 2018-10-19 | | 公開日 | 2018-11-21 | | 最終更新日 | 2019-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
2YBB
 
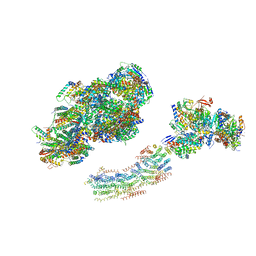 | | Fitted model for bovine mitochondrial supercomplex I1III2IV1 by single particle cryo-EM (EMD-1876) | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, CARDIOLIPIN, ... | | 著者 | Althoff, T, Mills, D.J, Popot, J.-L, Kuehlbrandt, W. | | 登録日 | 2011-03-02 | | 公開日 | 2011-10-19 | | 最終更新日 | 2019-10-23 | | 実験手法 | ELECTRON MICROSCOPY (19 Å) | | 主引用文献 | Arrangement of Electron Transport Chain Components in Bovine Mitochondrial Supercomplex I(1)III(2)Iv(1).
Embo J., 30, 2011
|
|
6MZC
 
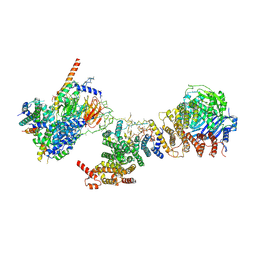 | | Human TFIID BC core | | 分子名称: | Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 12, Transcription initiation factor TFIID subunit 2, ... | | 著者 | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | 登録日 | 2018-11-05 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
4NO4
 
 | |
6N3P
 
 | | Crosslinked AcpP=FabZ complex from E. coli Type II FAS | | 分子名称: | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ, Acyl carrier protein, N~3~-{(2R)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-N-(3-{[(1Z)-pent-1-en-1-yl]sulfonyl}propyl)-beta-alaninamide | | 著者 | Smith, J.L, Dodge, G.J. | | 登録日 | 2018-11-15 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and dynamical rationale for fatty acid unsaturation inEscherichia coli.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
