2W1Q
 
 | |
2WDB
 
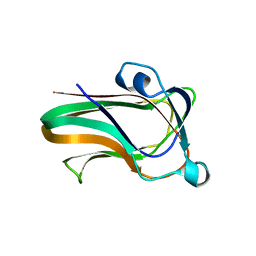 | | A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,2) mannose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, CALCIUM ION, HYALURONOGLUCOSAMINIDASE | | 著者 | Ficko-Blean, E, Boraston, A.B. | | 登録日 | 2009-03-23 | | 公開日 | 2009-05-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | N-Acetylglucosamine Recognition by a Family 32 Carbohydrate-Binding Module from Clostridium Perfringens Nagh.
J.Mol.Biol., 390, 2009
|
|
2W1S
 
 | |
7TDM
 
 | |
7TDN
 
 | |
2J1A
 
 | |
4D70
 
 | | Structural, biophysical and biochemical analyses of a Clostridium perfringens Sortase D5 transpeptidase | | 分子名称: | SORTASE FAMILY PROTEIN | | 著者 | Suryadinata, R, Seabrook, S, Adams, T.E, Nuttall, S.D, Peat, T.S. | | 登録日 | 2014-11-19 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural and Biochemical Analyses of a Clostridium Perfringens Sortase D Transpeptidase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3UCA
 
 | | Crystal structure of isoprenoid synthase (target EFI-501974) from clostridium perfringens | | 分子名称: | Geranyltranstransferase | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2011-10-26 | | 公開日 | 2011-11-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2WGV
 
 | | Crystal structure of the OXA-10 V117T mutant at pH 6.5 inhibited by a chloride ion | | 分子名称: | BETA-LACTAMASE OXA-10, CHLORIDE ION, CITRIC ACID, ... | | 著者 | Vercheval, L, Kerff, F, Bauvois, C, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | 登録日 | 2009-04-27 | | 公開日 | 2010-05-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post- Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
4UTT
 
 | | Structural characterisation of NanE, ManNac6P C2 epimerase, from Clostridium perfingens | | 分子名称: | ACETATE ION, CHLORIDE ION, PUTATIVE N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE | | 著者 | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | 登録日 | 2014-07-23 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway.
J.Biol.Chem., 289, 2014
|
|
6N68
 
 | |
4UTU
 
 | | Structural and biochemical characterization of the N- acetylmannosamine-6-phosphate 2-epimerase from Clostridium perfringens | | 分子名称: | CHLORIDE ION, N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE, N-acetylmannosamine-6-phosphate | | 著者 | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | 登録日 | 2014-07-23 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
J.Biol.Chem., 289, 2014
|
|
4FP9
 
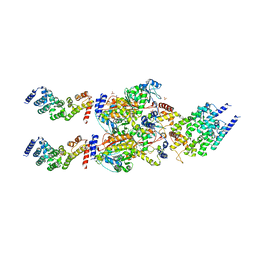 | | Human MTERF4-NSUN4 protein complex | | 分子名称: | S-ADENOSYLMETHIONINE, SULFATE ION, mTERF domain-containing protein 2, ... | | 著者 | Spahr, H, Hallberg, B.M. | | 登録日 | 2012-06-21 | | 公開日 | 2012-09-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of the human MTERF4-NSUN4 protein complex that regulates mitochondrial ribosome biogenesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8OVR
 
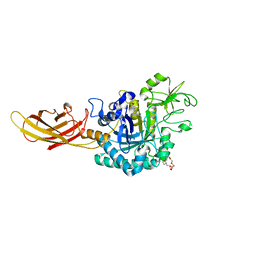 | |
8OWF
 
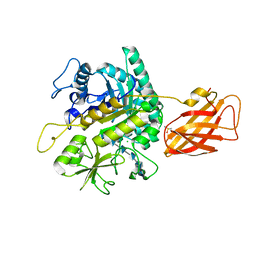 | | Clostridium perfringens chitinase CP4_3455 with chitosan | | 分子名称: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Chitodextrinase, ... | | 著者 | Bloch, Y, Savvides, S.N. | | 登録日 | 2023-04-27 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Clostridium perfringens chitinase CP4_3455 with chitosan
To Be Published
|
|
3L2B
 
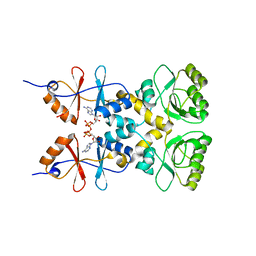 | | Crystal structure of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with activator, diadenosine tetraphosphate | | 分子名称: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Probable manganase-dependent inorganic pyrophosphatase | | 著者 | Tuominen, H, Salminen, A, Oksanen, E, Jamsen, J, Heikkila, O, Lehtio, L, Magretova, N.N, Goldman, A, Baykov, A.A, Lahti, R. | | 登録日 | 2009-12-15 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Crystal Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridiumperfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate.
J.Mol.Biol., 2010
|
|
3L31
 
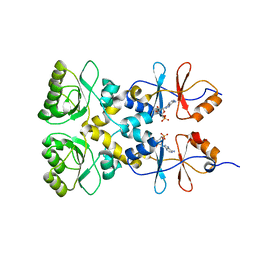 | | Crystal structure of the CBS and DRTGG domains of the regulatory region of Clostridium perfringens pyrophosphatase complexed with the inhibitor, AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, Probable manganase-dependent inorganic pyrophosphatase | | 著者 | Tuominen, H, Salminen, A, Oksanen, E, Jamsen, J, Heikkila, O, Lehtio, L, Magretova, N.N, Goldman, A, Baykov, A.A, Lahti, R. | | 登録日 | 2009-12-16 | | 公開日 | 2010-04-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of the CBS and DRTGG Domains of the Regulatory Region of Clostridiumperfringens Pyrophosphatase Complexed with the Inhibitor, AMP, and Activator, Diadenosine Tetraphosphate.
J.Mol.Biol., 2010
|
|
4RG2
 
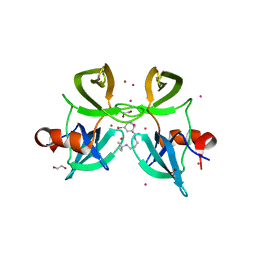 | | Tudor Domain of Tumor suppressor p53BP1 with small molecule ligand | | 分子名称: | 1,2-ETHANEDIOL, 3-bromo-N-[3-(tert-butylamino)propyl]benzamide, Tumor suppressor p53-binding protein 1, ... | | 著者 | Dong, A, Mader, P, James, L, Perfetti, M, Tempel, W, Frye, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | 登録日 | 2014-09-29 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Identification of a fragment-like small molecule ligand for the methyl-lysine binding protein, 53BP1.
ACS Chem. Biol., 10, 2015
|
|
1ETH
 
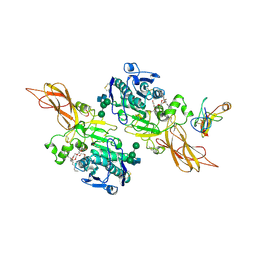 | | TRIACYLGLYCEROL LIPASE/COLIPASE COMPLEX | | 分子名称: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | 著者 | Hermoso, J, Pignol, D, Kerfelec, B, Crenon, I, Chapus, C, Fontecilla-Camps, J.C. | | 登録日 | 1995-09-13 | | 公開日 | 1996-12-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Lipase activation by nonionic detergents. The crystal structure of the porcine lipase-colipase-tetraethylene glycol monooctyl ether complex.
J.Biol.Chem., 271, 1996
|
|
7F5I
 
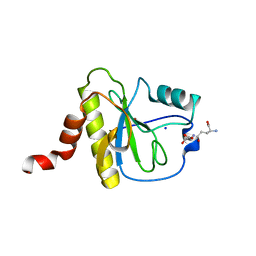 | | X-ray structure of Clostridium perfringens-specific amidase endolysin | | 分子名称: | GLUTAMIC ACID, SODIUM ION, ZINC ION, ... | | 著者 | Kamitori, S, Tamai, E. | | 登録日 | 2021-06-22 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and biochemical characterization of the Clostridium perfringens-specific Zn 2+ -dependent amidase endolysin, Psa, catalytic domain.
Biochem.Biophys.Res.Commun., 576, 2021
|
|
7O56
 
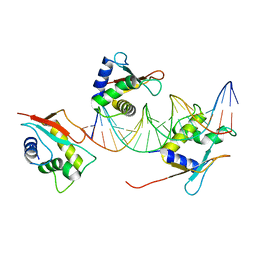 | | X-ray Structure of Interferon Regulatory Factor 4 DNA binding domain bound to an interferon-stimulated response element solved by Phosphorus and Sulphur SAD methods | | 分子名称: | DNA (5'-D(P*AP*AP*TP*AP*AP*AP*AP*GP*AP*AP*AP*CP*CP*GP*AP*AP*AP*GP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*AP*CP*TP*TP*TP*CP*GP*GP*TP*TP*TP*CP*TP*TP*TP*TP*AP*T)-3'), Interferon regulatory factor 4 | | 著者 | El Omari, K, Agnarelli, A, Duman, R, Wagner, A, Mancini, E.J. | | 登録日 | 2021-04-07 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Phosphorus and sulfur SAD phasing of the nucleic acid-bound DNA-binding domain of interferon regulatory factor 4.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6RQK
 
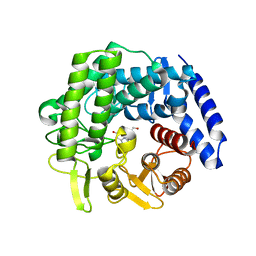 | | Crystal structure of GH125 1,6-alpha-mannosidase from Clostridium perfringens in complex with mannoimidazole | | 分子名称: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, Alpha-1,6-mannosidase | | 著者 | Males, A, Davies, G.J. | | 登録日 | 2019-05-16 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Distortion of mannoimidazole supports a B2,5boat transition state for the family GH125 alpha-1,6-mannosidase from Clostridium perfringens.
Org.Biomol.Chem., 17, 2019
|
|
5WQW
 
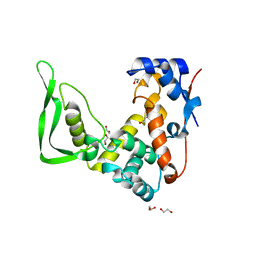 | | X-ray structure of catalytic domain of autolysin from Clostridium perfringens | | 分子名称: | 1,2-ETHANEDIOL, N-acetylglucosaminidase | | 著者 | Tamai, E, Sekiya, H, Goda, E, Makihata, N, Maki, J, Yoshida, H, Kamitori, S. | | 登録日 | 2016-11-29 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural and biochemical characterization of the Clostridium perfringens autolysin catalytic domain
FEBS Lett., 591, 2017
|
|
7D6P
 
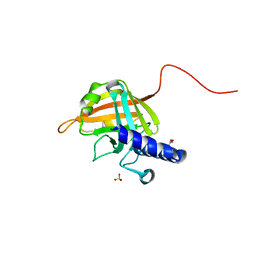 | |
7D6T
 
 | |
