1UM8
 
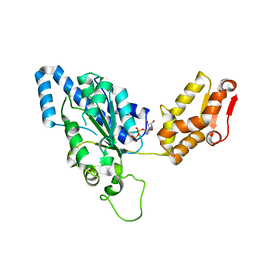 | | Crystal structure of helicobacter pylori ClpX | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit clpX | | 著者 | Kim, D.Y, Kim, K.K. | | 登録日 | 2003-09-25 | | 公開日 | 2003-12-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of ClpX Molecular Chaperone from Helicobacter pylori
J.Biol.Chem., 278, 2003
|
|
3LVG
 
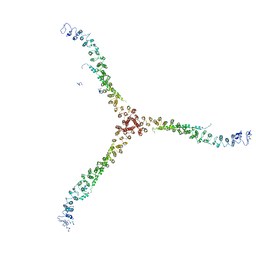 | | Crystal structure of a clathrin heavy chain and clathrin light chain complex | | 分子名称: | Clathrin heavy chain 1, Clathrin light chain B | | 著者 | Wilbur, J.D, Hwang, P.K, Ybe, J.A, Lane, M, Sellers, B.D, Jacobson, M.P, Fletterick, R.J, Brodsky, F.M. | | 登録日 | 2010-02-20 | | 公開日 | 2010-06-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (7.94 Å) | | 主引用文献 | Conformation switching of clathrin light chain regulates clathrin lattice assembly.
Dev.Cell, 18, 2010
|
|
7FJM
 
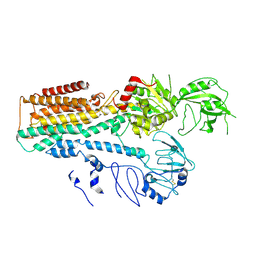 | | Cryo EM structure of lysosomal ATPase | | 分子名称: | Polyamine-transporting ATPase 13A2 | | 著者 | Zhang, S.S. | | 登録日 | 2021-08-04 | | 公開日 | 2023-03-08 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2.
Cell Discov, 7, 2021
|
|
4FGV
 
 | |
7FJP
 
 | | Cryo EM structure of lysosomal ATPase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Zhang, S.S. | | 登録日 | 2021-08-04 | | 公開日 | 2023-03-08 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2.
Cell Discov, 7, 2021
|
|
5IC5
 
 | | Bacteriophytochrome response regulator RtBRR | | 分子名称: | CACODYLATE ION, Candidate response regulator, CheY, ... | | 著者 | Baker, A.W, Satyshur, K.A, Forest, K.T. | | 登録日 | 2016-02-22 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Arm-in-Arm Response Regulator Dimers Promote Intermolecular Signal Transduction.
J.Bacteriol., 198, 2016
|
|
6LDR
 
 | |
6LDS
 
 | |
6LDT
 
 | |
7X3X
 
 | | Cryo-EM structure of N1 nucleosome-RA | | 分子名称: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Lifei, L, Kangjing, C, Chen, Z. | | 登録日 | 2022-03-01 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1GSN
 
 | | HUMAN GLUTATHIONE REDUCTASE MODIFIED BY DINITROSOGLUTATHIONE | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, GLUTATHIONE REDUCTASE, ... | | 著者 | Becker, K, Savvides, S.N, Keese, M, Schirmer, R.H, Karplus, P.A. | | 登録日 | 1998-02-21 | | 公開日 | 1998-05-27 | | 最終更新日 | 2011-12-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Enzyme inactivation through sulfhydryl oxidation by physiologic NO-carriers.
Nat.Struct.Biol., 5, 1998
|
|
2GNV
 
 | |
4HZK
 
 | |
8I74
 
 | |
8I75
 
 | |
8I76
 
 | |
7COW
 
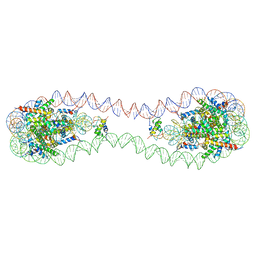 | | 353 bp di-nucleosome harboring cohesive DNA termini with linker histone H1.0 | | 分子名称: | CALCIUM ION, CHLORIDE ION, DNA (353-MER), ... | | 著者 | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | 登録日 | 2020-08-05 | | 公開日 | 2021-08-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
3DTU
 
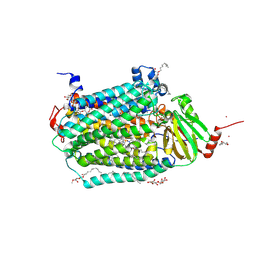 | | Catalytic core subunits (I and II) of cytochrome c oxidase from Rhodobacter sphaeroides complexed with deoxycholic acid | | 分子名称: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, CADMIUM ION, CALCIUM ION, ... | | 著者 | Qin, L, Mills, D.A, Buhrow, L, Hiser, C, Ferguson-Miller, S. | | 登録日 | 2008-07-15 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | A conserved steroid binding site in cytochrome C oxidase.
Biochemistry, 47, 2008
|
|
5TN2
 
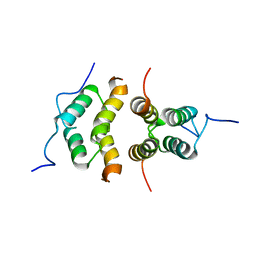 | | Solution Structure of the C-terminal multimerization domain of the master biofilm-regulator SinR from Bacillus subtilis | | 分子名称: | HTH-type transcriptional regulator SinR | | 著者 | Draughn, G.L, Bobay, B.G, Stowe, S.D, Thompson, R.J, Cavanagh, J. | | 登録日 | 2016-10-13 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Solution Structures and Interaction of SinR and SinI: Elucidating the Mechanism of Action of the Master Regulator Switch for Biofilm Formation in Bacillus subtilis.
J.Mol.Biol., 2019
|
|
5VAR
 
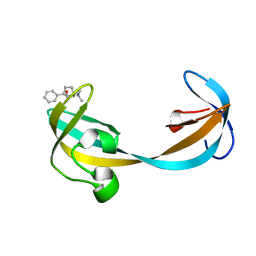 | |
7E1B
 
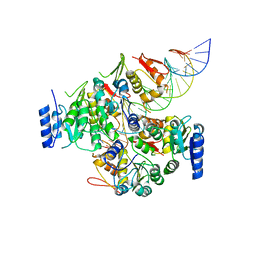 | | Crystal structure of VbrR-DNA complex | | 分子名称: | DNA (26-MER), DNA-binding response regulator | | 著者 | Hong, S, Zhang, X, Zhang, P. | | 登録日 | 2021-02-01 | | 公開日 | 2022-02-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (4.587 Å) | | 主引用文献 | Structural basis of phosphorylation-induced activation of the response regulator VbrR.
Acta Biochim.Biophys.Sin., 2023
|
|
8V5M
 
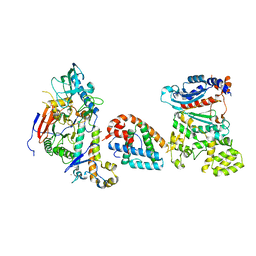 | | Tetramer core subcomplex (conformation 1) of Xenopus laevis DNA polymerase alpha-primase | | 分子名称: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | 著者 | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | 登録日 | 2023-11-30 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (9.22 Å) | | 主引用文献 | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5O
 
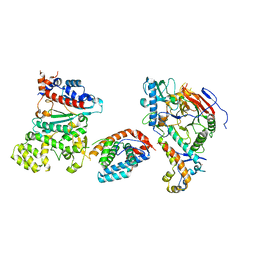 | | Tetramer core subcomplex (conformation 3) of Xenopus laevis DNA polymerase alpha-primase | | 分子名称: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | 著者 | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | 登録日 | 2023-11-30 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (8.99 Å) | | 主引用文献 | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5N
 
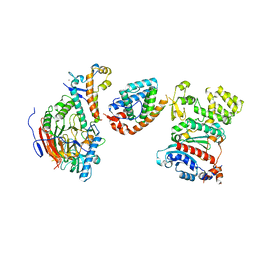 | | Tetramer core subcomplex (conformation 2) of Xenopus laevis DNA polymerase alpha-primase | | 分子名称: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | 著者 | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | 登録日 | 2023-11-30 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (8.56 Å) | | 主引用文献 | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7X3W
 
 | | Cryo-EM structure of ISW1-N1 nucleosome | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (146-MER), ... | | 著者 | Lifei, L, Kangjing, C, Chen, Z. | | 登録日 | 2022-03-01 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
