4HF0
 
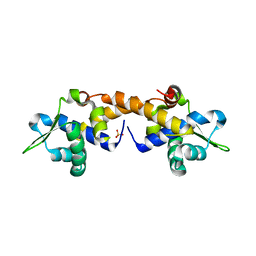 | | Crystal Structure of Apo IscR | | 分子名称: | HTH-type transcriptional regulator IscR, SULFATE ION | | 著者 | Rajagopalan, S.R, Phillips, K.J. | | 登録日 | 2012-10-04 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Studies of IscR reveal a unique mechanism for metal-dependent regulation of DNA binding specificity.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2YGK
 
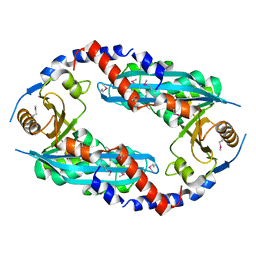 | | Crystal structure of the NurA nuclease from Sulfolobus solfataricus | | 分子名称: | MANGANESE (II) ION, NURA | | 著者 | Rzechorzek, N.J, Blackwood, J.K, Abrams, A.S, Maman, J.D, Robinson, N.P, Pellegrini, L. | | 登録日 | 2011-04-18 | | 公開日 | 2011-12-14 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and Functional Insights Into DNA-End Processing by the Archaeal Hera Helicase-Nura Nuclease Complex.
Nucleic Acids Res., 40, 2012
|
|
1J53
 
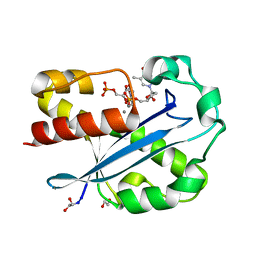 | | Structure of the N-terminal Exonuclease Domain of the Epsilon Subunit of E.coli DNA Polymerase III at pH 8.5 | | 分子名称: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | 著者 | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | 登録日 | 2002-01-22 | | 公開日 | 2002-10-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
366D
 
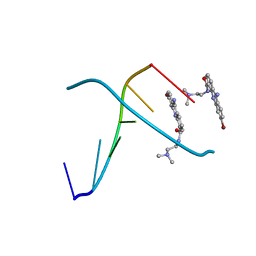 | | 1.3 A STRUCTURE DETERMINATION OF THE D(CG(5-BRU)ACG)2/6-BROMO-9-AMINO-DACA COMPLEX | | 分子名称: | 6-BROMO-9-AMINO-N-ETHYL(DIAMINOMETHYL)ACRIDINE-4-CARBOXAMIDE, DNA (5'-D(*CP*GP*(BRU)P*AP*CP*G)-3') | | 著者 | Todd, A.K, Adams, A, Thorpe, J.H, Denny, W.A, Cardin, C.J. | | 登録日 | 1997-12-19 | | 公開日 | 1999-04-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Major groove binding and 'DNA-induced' fit in the intercalation of a derivative of the mixed topoisomerase I/II poison N-(2-(dimethylamino)ethyl)acridine-4-carboxamide (DACA) into DNA: X-ray structure complexed to d(CG(5-BrU)ACG)2 at 1.3-A resolution.
J.Med.Chem., 42, 1999
|
|
4OFH
 
 | |
2IVO
 
 | | Structure of UP1 protein | | 分子名称: | TUNGSTATE(VI)ION, UP1 | | 著者 | Hecker, A, Leulliot, N, Graille, M, Dorlet, P, Quevillon-Cheruel, S, Ulryck, N, Van Tilbeurgh, H, Forterre, P. | | 登録日 | 2006-06-14 | | 公開日 | 2007-07-31 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | An Archaeal Orthologue of the Universal Protein Kae1 is an Iron Metalloprotein which Exhibits Atypical DNA-Binding Properties and Apurinic-Endonuclease Activity in Vitro.
Nucleic Acids Res., 35, 2007
|
|
3FLD
 
 | |
6B47
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF2 | | 分子名称: | Anti-CRISPR protein AcrF2, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | 著者 | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | 登録日 | 2017-09-25 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B48
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF10 | | 分子名称: | Anti-CRISPR protein AcrF10, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | 著者 | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | 登録日 | 2017-09-25 | | 公開日 | 2017-10-18 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
8EXH
 
 | | Agrobacterium tumefaciens Tpilus | | 分子名称: | (14S,17R)-20-amino-17-hydroxy-11,17-dioxo-12,16,18-trioxa-17lambda~5~-phosphaicosan-14-yl tetradecanoate, Protein virB2 | | 著者 | Beltran, L.C, Egelman, E.H. | | 登録日 | 2022-10-25 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-09-25 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery
Nat Commun, 14, 2023
|
|
8EJO
 
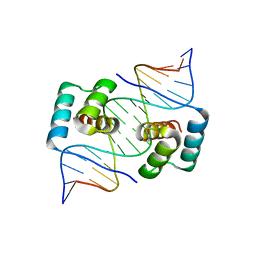 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | 著者 | Yin, L, Shi, K, Aihara, H. | | 登録日 | 2022-09-18 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
8EJP
 
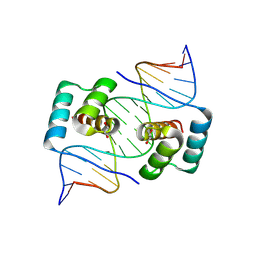 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA containing 5-Bromouracil | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | 著者 | Yin, L, Shi, K, Aihara, H. | | 登録日 | 2022-09-18 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (2.174 Å) | | 主引用文献 | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
4ODS
 
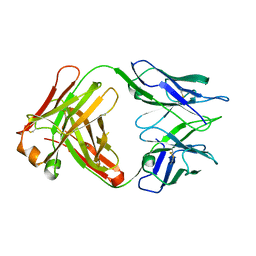 | |
4ODV
 
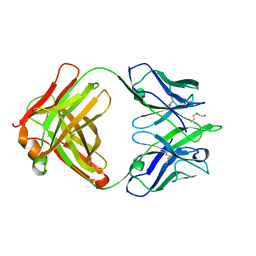 | |
6FN0
 
 | | The animal-like Cryptochrome from Chlamydomonas reinhardtii in complex with 6-4 DNA | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Cryptochrome photoreceptor, ... | | 著者 | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Yamamoto, J, Mittag, M, Essen, L.-O. | | 登録日 | 2018-02-02 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii.
Nucleic Acids Res., 46, 2018
|
|
6S03
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor I39LT379 | | 分子名称: | 4-[[4-[5,5-dimethyl-2-(6-methylpyridin-2-yl)-4,6-dihydropyrrolo[1,2-b]pyrazol-3-yl]pyridin-2-yl]amino]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | 著者 | Kugler, M, Brynda, J, Rezacova, P. | | 登録日 | 2019-06-13 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA).
SLAS Discov, 25, 2020
|
|
8VTV
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolone MCX-91, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.55A resolution | | 分子名称: | 1-cyclopropyl-7-{7-[({[(3aR,4R,7R,8S,9S,10R,11R,13R,14E,15S,15aR)-10-{[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy}-4-ethyl-11-methoxy-14-(methoxyimino)-3a,7,9,11,13,15-hexamethyl-2,6-dioxododecahydro-2H,4H-[1,3]dioxolo[4,5-c]oxacyclotetradecin-8-yl]oxy}carbonyl)amino]hept-1-yn-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | 著者 | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | 登録日 | 2024-01-27 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 2024
|
|
2WIW
 
 | | Crystal structures of Holliday junction resolvases from Archaeoglobus fulgidus bound to DNA substrate | | 分子名称: | 5'-D(*DC*DG*DG*DA*DT*DA*DT*DC*DC*DGP)-3', GLYCEROL, HEXANE-1,6-DIOL, ... | | 著者 | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | 登録日 | 2009-05-18 | | 公開日 | 2009-05-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of Holliday Junction Resolvases from Archaeoglobus Fulgidus Bound to DNA Substrate
To be Published
|
|
5NOC
 
 | |
5NVR
 
 | |
8VTU
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolone MCX-66, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.40A resolution | | 分子名称: | 1-cyclopropyl-7-[(4-{[3-({(3aR,4R,7R,8S,9S,10R,11R,13R,14E,15S,15aR)-10-{[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy}-4-ethyl-11-methoxy-3a,7,9,11,13,15-hexamethyl-14-[({3-[5-(methylcarbamoyl)pyridin-3-yl]prop-2-yn-1-yl}oxy)imino]-2,6-dioxododecahydro-2H,4H-[1,3]dioxolo[4,5-c]oxacyclotetradecin-8-yl}oxy)-3-oxopropyl]amino}butyl)amino]-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | 著者 | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | 登録日 | 2024-01-27 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 2024
|
|
8VTX
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with macrolone MCX-128, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.40A resolution | | 分子名称: | 1-cyclopropyl-7-(4-{4-[({[(3aR,4R,7R,8S,9S,10R,11R,13R,14E,15S,15aR)-10-{[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy}-4-ethyl-11-methoxy-14-(methoxyimino)-3a,7,9,11,13,15-hexamethyl-2,6-dioxododecahydro-2H,4H-[1,3]dioxolo[4,5-c]oxacyclotetradecin-8-yl]oxy}carbonyl)amino]butyl}piperazin-1-yl)-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | 著者 | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | 登録日 | 2024-01-27 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 2024
|
|
4ODT
 
 | |
8GJA
 
 | | RAD51C-XRCC3 structure | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, RAD51C, ... | | 著者 | Arvai, A.S, Tainer, J.A, Williams, G, Longo, M.A. | | 登録日 | 2023-03-15 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles.
Nat Commun, 14, 2023
|
|
8OJ1
 
 | |
