7BV9
 
 | | The NMR structure of the BEN domain from human NAC1 | | 分子名称: | Nucleus accumbens-associated protein 1 | | 著者 | Nagata, T, Kobayashi, N, Nakayama, N, Obayashi, E, Urano, T. | | 登録日 | 2020-04-09 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Nucleus Accumbens-Associated Protein 1 Binds DNA Directly through the BEN Domain in a Sequence-Specific Manner.
Biomedicines, 8, 2020
|
|
6K07
 
 | | Crystal structure of REV7(R124A) in complex with a Shieldin3 fragment | | 分子名称: | Mitotic spindle assembly checkpoint protein MAD2B, SULFATE ION, Shieldin complex subunit 3 | | 著者 | Zhang, F, Dai, Y. | | 登録日 | 2019-05-05 | | 公開日 | 2019-12-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural basis for shieldin complex subunit 3-mediated recruitment of the checkpoint protein REV7 during DNA double-strand break repair.
J.Biol.Chem., 295, 2020
|
|
2OWL
 
 | | Crystal structure of E. coli RdgC | | 分子名称: | CALCIUM ION, Recombination-associated protein rdgC | | 著者 | Briggs, G.S, McEwan, P.A, Yu, J, Moore, T, Emsley, J, Lloyd, R.G. | | 登録日 | 2007-02-16 | | 公開日 | 2007-03-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Ring Structure of the Escherichia coli DNA-binding Protein RdgC Associated with Recombination and Replication Fork Repair.
J.Biol.Chem., 282, 2007
|
|
2K4J
 
 | |
2L4M
 
 | |
2QBY
 
 | | Crystal structure of a heterodimer of Cdc6/Orc1 initiators bound to origin DNA (from S. solfataricus) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 6 homolog 1, Cell division control protein 6 homolog 3, ... | | 著者 | Cunningham Dueber, E.L, Corn, J.E, Bell, S.D, Berger, J.M. | | 登録日 | 2007-06-18 | | 公開日 | 2007-09-11 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex.
Science, 317, 2007
|
|
6RYK
 
 | |
2DIM
 
 | | Solution structure of the Myb_DNA-binding domain of human Cell division cycle 5-like protein | | 分子名称: | Cell division cycle 5-like protein | | 著者 | Yoneyama, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-30 | | 公開日 | 2007-04-03 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Myb_DNA-binding domain of human Cell division cycle 5-like protein
To be Published
|
|
2DIN
 
 | | Solution structure of the Myb_DNA-binding domain of human Cell division cycle 5-like protein | | 分子名称: | Cell division cycle 5-like protein | | 著者 | Yoneyama, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-30 | | 公開日 | 2007-04-03 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Myb_DNA-binding domain of human Cell division cycle 5-like protein
To be Published
|
|
8OJ1
 
 | |
8EJO
 
 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | 著者 | Yin, L.L, Shi, K, Aihara, H. | | 登録日 | 2022-09-18 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
8EJP
 
 | | Crystal structure of the homeodomain of Platypus sDUX in complex with DNA containing 5-Bromouracil | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), ... | | 著者 | Yin, L.L, Shi, K, Aihara, H. | | 登録日 | 2022-09-18 | | 公開日 | 2023-09-20 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.174 Å) | | 主引用文献 | Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein.
Iscience, 26, 2023
|
|
6ANV
 
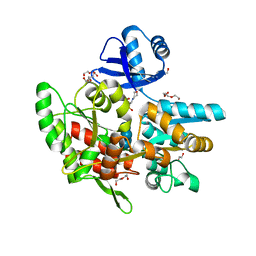 | | Crystal structure of anti-CRISPR protein AcrF1 | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Yang, H, Patel, D.J. | | 登録日 | 2017-08-14 | | 公開日 | 2017-10-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.265 Å) | | 主引用文献 | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
3FCK
 
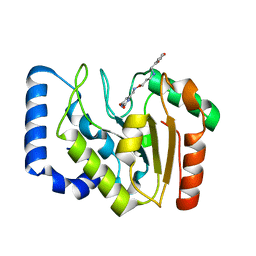 | | Complex of UNG2 and a fragment-based design inhibitor | | 分子名称: | 3-({[3-({[(1E)-(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)methylidene]amino}oxy)propyl]amino}methyl)benzoic acid, Uracil-DNA glycosylase | | 著者 | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | 登録日 | 2008-11-21 | | 公開日 | 2009-04-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
7ECW
 
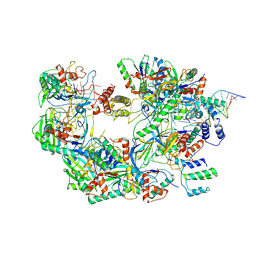 | | The Csy-AcrIF14-dsDNA complex | | 分子名称: | 54-MER DNA, AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, ... | | 著者 | Zhang, L.X, Feng, Y. | | 登録日 | 2021-03-13 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
1AE9
 
 | |
1F0O
 
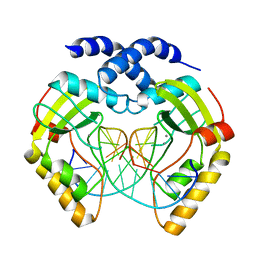 | |
378D
 
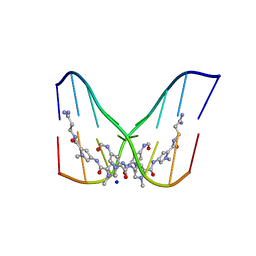 | | STRUCTURE OF THE SIDE-BY-SIDE BINDING OF DISTAMYCIN TO DNA | | 分子名称: | DISTAMYCIN A, DNA (5'-D(*GP*TP*AP*TP*AP*TP*AP*C)-3'), SODIUM ION | | 著者 | Mitra, S.N, Wahl, M.C, Sundaralingam, M. | | 登録日 | 1998-01-28 | | 公開日 | 1999-03-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of the side-by-side binding of distamycin to d(GTATATAC)2.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2H7G
 
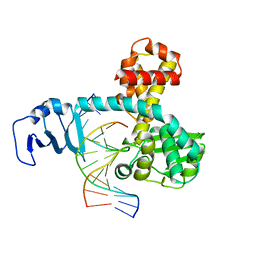 | | Structure of variola topoisomerase non-covalently bound to DNA | | 分子名称: | 5'-D(*TP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*AP*CP*A)-3', 5'-D(*TP*TP*GP*TP*CP*GP*CP*CP*CP*TP*TP*A)-3', DNA topoisomerase 1 | | 著者 | Perry, K, Hwang, Y, Bushman, F.D, Van Duyne, G.D. | | 登録日 | 2006-06-02 | | 公開日 | 2006-08-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for specificity in the poxvirus topoisomerase.
Mol.Cell, 23, 2006
|
|
7QV1
 
 | |
7QV2
 
 | |
3FCI
 
 | | Complex of UNG2 and a fragment-based designed inhibitor | | 分子名称: | 3-{(E)-[(3-{[(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)methyl]amino}propoxy)imino]methyl}benzoic acid, SODIUM ION, THIOCYANATE ION, ... | | 著者 | Bianchet, M.A, Chung, S, Parker, J.B, Amzel, L.M, Stivers, J.T. | | 登録日 | 2008-11-21 | | 公開日 | 2009-04-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Impact of linker strain and flexibility in the design of a fragment-based inhibitor
Nat.Chem.Biol., 5, 2009
|
|
3MIS
 
 | | I-MsoI re-designed for altered DNA cleavage specificity (-8G) | | 分子名称: | CALCIUM ION, DNA (5'-D(*CP*GP*GP*AP*GP*CP*TP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*GP*TP*CP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*GP*GP*AP*CP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*AP*GP*CP*TP*CP*CP*G)-3'), ... | | 著者 | Taylor, G.K, Stoddard, B.L. | | 登録日 | 2010-04-12 | | 公開日 | 2010-05-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs.
Nucleic Acids Res., 38, 2010
|
|
4ATI
 
 | | MITF:M-box complex | | 分子名称: | 5'-D(*AP*GP*GP*GP*TP*CP*AP*TP*GP*TP*GP*CP*TP*AP*AP*C)-3', 5'-D(*GP*TP*TP*AP*GP*CP*AP*CP*AP*TP*GP*AP*CP*CP*CP*T)-3', MICROPHTHALMIA-ASSOCIATED TRANSCRIPTION FACTOR | | 著者 | Pogenberg, V, Deineko, V, Wilmanns, M. | | 登録日 | 2012-05-08 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Restricted Leucine Zipper Dimerization and Specificity of DNA Recognition of the Melanocyte Master Regulator Mitf
Genes Dev., 26, 2012
|
|
1EDR
 
 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO6AATTCGCG) AT 1.6 ANGSTROM | | 分子名称: | 5'-D(*CP*GP*CP*GP*(A47)AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION, SPERMINE | | 著者 | Chatake, T, Hikima, T, Ono, A, Ueno, Y, Matsuda, A, Takenaka, A. | | 登録日 | 2000-01-28 | | 公開日 | 2000-02-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystallographic studies on damaged DNAs. II. N(6)-methoxyadenine can present two alternate faces for Watson-Crick base-pairing, leading to pyrimidine transition mutagenesis.
J.Mol.Biol., 294, 1999
|
|
