2M93
 
 | |
8U4W
 
 | |
6BZG
 
 | |
5VAJ
 
 | | BhRNase H - amide-RNA/DNA complex | | 分子名称: | ACETATE ION, DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), GLYCEROL, ... | | 著者 | Pallan, P.S, Egli, M. | | 登録日 | 2017-03-27 | | 公開日 | 2017-09-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Amide linkages mimic phosphates in RNA interactions with proteins and are well tolerated in the guide strand of short interfering RNAs.
Nucleic Acids Res., 45, 2017
|
|
5A78
 
 | | Crystal structure of the homing endonuclease I-CvuI in complex with I- CreI target (C1221) in the presence of 2 mM Mg revealing DNA not cleaved | | 分子名称: | 24MER DNA, 5'-D(*TP*CP*AP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP *CP*GP*AP*CP*GP*TP*TP*TP*TP*GP*A)-3', DNA ENDONUCLEASE I-CVUI, ... | | 著者 | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | 登録日 | 2015-07-03 | | 公開日 | 2015-09-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
5YZY
 
 | | AtVAL1 B3 domain in complex with 13bp-DNA | | 分子名称: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*A)-3'), ... | | 著者 | Wu, B.X, Zhang, M.M. | | 登録日 | 2017-12-17 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structural insight into the role of VAL1 B3 domain for targeting to FLC locus in Arabidopsis thaliana.
Biochem. Biophys. Res. Commun., 2018
|
|
2IH5
 
 | |
7OT6
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND inhibitor RMC-282 | | 分子名称: | (R)-N-(1-(6-amino-9H-purin-9-yl)propan-2-yl)-N-(2-phosphonoethyl)glycine, DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), ... | | 著者 | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | 登録日 | 2021-06-09 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OTX
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-257 | | 分子名称: | (S)-2-((3-(6-amino-9H-purin-9-yl)propyl)amino)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | 登録日 | 2021-06-10 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OUT
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-264 | | 分子名称: | (S)-2-(3-(6-amino-9H-purin-9-yl)propoxy)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | 登録日 | 2021-06-13 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OTK
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-233 | | 分子名称: | (2~{R})-2-[2-(6-aminopurin-9-yl)ethylamino]-3-phosphono-propanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | 登録日 | 2021-06-10 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OTA
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-230 | | 分子名称: | ((2-(6-amino-9H-purin-9-yl)ethyl)-L-seryl)phosphoramidic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | 登録日 | 2021-06-09 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
6BZF
 
 | |
1RYU
 
 | | Solution Structure of the SWI1 ARID | | 分子名称: | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin subfamily F member 1 | | 著者 | Kim, S, Zhang, Z, Upchurch, S, Isern, N, Chen, Y. | | 登録日 | 2003-12-22 | | 公開日 | 2004-05-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and DNA-binding sites of the SWI1 AT-rich interaction domain (ARID) suggest determinants for sequence-specific DNA recognition.
J.Biol.Chem., 279, 2004
|
|
7OTN
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-247 | | 分子名称: | (S)-2-((2-(6-amino-9H-purin-9-yl)ethyl)amino)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | 登録日 | 2021-06-10 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OTZ
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-259 | | 分子名称: | (S)-2-(2-(6-amino-9H-purin-9-yl)ethoxy)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | 登録日 | 2021-06-10 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
1S7E
 
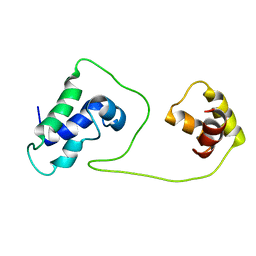 | | Solution structure of HNF-6 | | 分子名称: | Hepatocyte nuclear factor 6 | | 著者 | Liao, X, Sheng, W. | | 登録日 | 2004-01-29 | | 公開日 | 2004-12-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the hepatocyte nuclear factor 6alpha and its interaction with DNA.
J.Biol.Chem., 279, 2004
|
|
7O0N
 
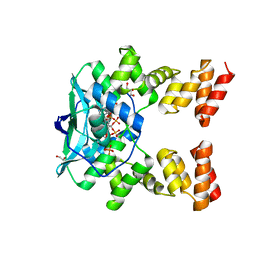 | |
6BHV
 
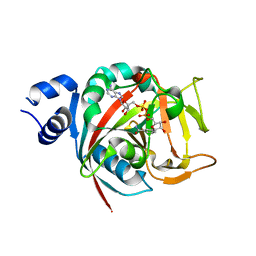 | | Human PARP-1 bound to NAD+ analog benzamide adenine dinucleotide (BAD) | | 分子名称: | Poly [ADP-ribose] polymerase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3S,4R,5S)-5-(3-carbamoylphenyl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name) | | 著者 | Pascal, J.M, Langelier, M.F. | | 登録日 | 2017-10-31 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | NAD+analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains.
Nat Commun, 9, 2018
|
|
4M32
 
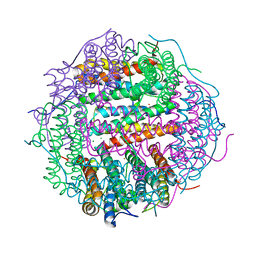 | | Crystal structure of gated-pore mutant D138N of second DNA-Binding protein under starvation from Mycobacterium smegmatis | | 分子名称: | CHLORIDE ION, FE (II) ION, MAGNESIUM ION, ... | | 著者 | Williams, S.M, Chandran, A.V, Vijayabaskar, M.S, Roy, S, Balaram, H, Vishveshwara, S, Vijayan, M, Chatterji, D. | | 登録日 | 2013-08-06 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | A histidine aspartate ionic lock gates the iron passage in miniferritins from Mycobacterium smegmatis
J.Biol.Chem., 289, 2014
|
|
6C5D
 
 | |
8KAG
 
 | |
3M7K
 
 | |
5YZZ
 
 | | AtVAL1 B3 domain in complex with 13bp-DNA | | 分子名称: | B3 domain-containing transcription repressor VAL1, DNA (5'-D(*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*A)-3') | | 著者 | Wu, B.X, Zhang, M.M. | | 登録日 | 2017-12-17 | | 公開日 | 2018-05-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structural insight into the role of VAL1 B3 domain for targeting to FLC locus in Arabidopsis thaliana.
Biochem. Biophys. Res. Commun., 2018
|
|
4M94
 
 | |
