4U4M
 
 | | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate | | 分子名称: | 1,2-ETHANEDIOL, PYRUVIC ACID, UREA, ... | | 著者 | Manoj Kumar, P, Bhaskar, V, Manicka, S, Krishnaswamy, S. | | 登録日 | 2014-07-24 | | 公開日 | 2015-07-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate
To be published
|
|
4U94
 
 | | Structure of mycobacterial maltokinase, the missing link in the essential GlgE-pathway | | 分子名称: | MAGNESIUM ION, Maltokinase | | 著者 | Fraga, J, Empadinhas, N, Pereira, P.J.B, Macedo-Ribeiro, S. | | 登録日 | 2014-08-05 | | 公開日 | 2015-02-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.473 Å) | | 主引用文献 | Structure of mycobacterial maltokinase, the missing link in the essential GlgE-pathway.
Sci Rep, 5, 2015
|
|
4U9P
 
 | | Structure of the methanofuran/methanopterin biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii | | 分子名称: | GLYCEROL, UPF0264 protein MJ1099 | | 著者 | Bobik, T.A, Morales, E, Shin, A, Cascio, D, Sawaya, M.R, Arbing, M, Rasche, M.E. | | 登録日 | 2014-08-06 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of the methanofuran/methanopterin-biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4U9Z
 
 | |
7D58
 
 | | cryo-EM structure of human RNA polymerase III in elongating state | | 分子名称: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | 著者 | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | 登録日 | 2020-09-25 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D59
 
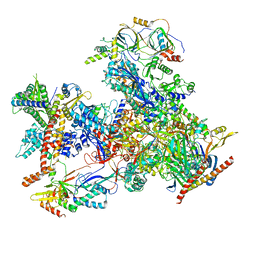 | | cryo-EM structure of human RNA polymerase III in apo state | | 分子名称: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | 著者 | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | 登録日 | 2020-09-25 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7DLA
 
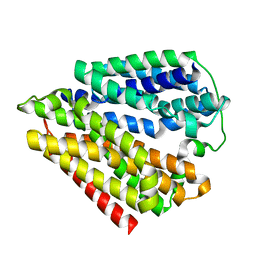 | |
7DAC
 
 | | Human RIPK3 amyloid fibril revealed by solid-state NMR | | 分子名称: | Receptor-interacting serine/threonine-protein kinase 3 | | 著者 | Wu, X.L, Zhang, J, Dong, X.Q, Liu, J, Li, B, Hu, H, Wang, J, Wang, H.Y, Lu, J.X. | | 登録日 | 2020-10-16 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DA4
 
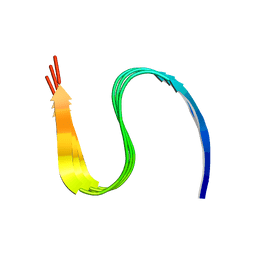 | | Cryo-EM structure of amyloid fibril formed by human RIPK3 | | 分子名称: | Receptor-interacting serine/threonine-protein kinase 3 | | 著者 | Zhao, K, Ma, Y.Y, Sun, Y.P, Li, D, Liu, C. | | 登録日 | 2020-10-14 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.24 Å) | | 主引用文献 | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DD5
 
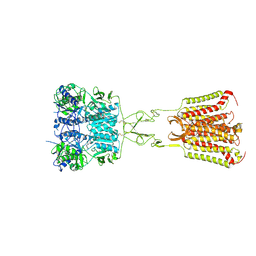 | | Structure of Calcium-Sensing Receptor in complex with NPS-2143 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, ... | | 著者 | Wen, T.L, Yang, X, Shen, Y.Q. | | 登録日 | 2020-10-27 | | 公開日 | 2021-06-16 | | 最終更新日 | 2021-06-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor.
Sci Adv, 7, 2021
|
|
7DD7
 
 | | Structure of Calcium-Sensing Receptor in complex with Evocalcet | | 分子名称: | 2-[4-[(3S)-3-[[(1R)-1-naphthalen-1-ylethyl]amino]pyrrolidin-1-yl]phenyl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wen, T.L, Yang, X, Shen, Y.Q. | | 登録日 | 2020-10-27 | | 公開日 | 2021-06-16 | | 最終更新日 | 2021-06-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor.
Sci Adv, 7, 2021
|
|
7DD6
 
 | | Structure of Ca2+/L-Trp-bonnd Calcium-Sensing Receptor in active state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Wen, T.L, Yang, X, Shen, Y.Q. | | 登録日 | 2020-10-27 | | 公開日 | 2021-06-16 | | 最終更新日 | 2021-06-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor.
Sci Adv, 7, 2021
|
|
2HUG
 
 | | 3D Solution Structure of the Chromo-2 Domain of cpSRP43 complexed with cpSRP54 peptide | | 分子名称: | Signal recognition particle 43 kDa protein, chloroplast, Signal recognition particle 54 kDa protein | | 著者 | Kathir, K.M, Vaithiyalingam, S, Henry, R, Thallapuranam, S.K.K. | | 登録日 | 2006-07-26 | | 公開日 | 2007-09-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Assembly of chloroplast signal recognition particle involves structural rearrangement in cpSRP43.
J.Mol.Biol., 381, 2008
|
|
2RUH
 
 | | Chemical Shift Assignments for MIP and MDM2 in bound state | | 分子名称: | E3 ubiquitin-protein ligase Mdm2 | | 著者 | Nagata, T, Shirakawa, K, Kobayashi, N, Shiheido, H, Horisawa, K, Katahira, M, Doi, N, Yanagawa, H. | | 登録日 | 2014-06-03 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for Inhibition of the MDM2:p53 Interaction by an Optimized MDM2-Binding Peptide Selected with mRNA Display
Plos One, 9, 2014
|
|
7DFB
 
 | | Crystal of Arrestin2-V2Rpp-6-7-Fab30 complex | | 分子名称: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | 著者 | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | 登録日 | 2020-11-06 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.28 Å) | | 主引用文献 | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DF9
 
 | | Crystal of Arrestin2-V2Rpp-1-Fab30 complex | | 分子名称: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | 著者 | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | 登録日 | 2020-11-06 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.17 Å) | | 主引用文献 | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DFA
 
 | | Crystal of Arrestin2-V2Rpp-4-Fab30 complex | | 分子名称: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | 著者 | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | 登録日 | 2020-11-06 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7DFC
 
 | | Crystal of Arrestin2-V2Rpp-3-Fab30 complex | | 分子名称: | Beta-arrestin-1, FAB30 HEAVY CHAIN, FAB30 LIGHT CHAIN, ... | | 著者 | Sun, J.P, Yu, X, Xiao, P, He, Q.T, Lin, J.Y, Zhu, Z.L. | | 登録日 | 2020-11-06 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Structural studies of phosphorylation-dependent interactions between the V2R receptor and arrestin-2.
Nat Commun, 12, 2021
|
|
7CYM
 
 | | Crystal structure of LI-Cadherin EC1-4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Caaveiro, J.M.M, Yui, A, Tsumoto, K. | | 登録日 | 2020-09-03 | | 公開日 | 2021-08-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Mechanism of dimerization and structural features of human LI-cadherin.
J.Biol.Chem., 297, 2021
|
|
7DP1
 
 | |
7DP0
 
 | |
7DP2
 
 | |
2HG0
 
 | | Structure of the West Nile Virus envelope glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein | | 著者 | Nybakken, G.E, Nelson, C.A, Chen, B.R, Diamond, M.S, Fremont, D.H. | | 登録日 | 2006-06-26 | | 公開日 | 2006-11-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of the West Nile virus envelope glycoprotein.
J.Virol., 80, 2006
|
|
7DNU
 
 | | mRNA-decapping enzyme g5Rp with inhibitor insp6 complex | | 分子名称: | INOSITOL HEXAKISPHOSPHATE, mRNA-decapping protein g5R | | 著者 | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | 登録日 | 2020-12-10 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.245 Å) | | 主引用文献 | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
7DNT
 
 | | mRNA-decapping enzyme g5Rp | | 分子名称: | mRNA-decapping protein g5R | | 著者 | Yang, Y, Chen, C, Li, L, Li, X.H, Su, D. | | 登録日 | 2020-12-10 | | 公開日 | 2022-03-09 | | 最終更新日 | 2022-12-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
J.Virol., 96, 2022
|
|
