6S58
 
 | | AvaII restriction endonuclease in the absence of nucleic acids | | 分子名称: | CALCIUM ION, Type II site-specific deoxyribonuclease, UNKNOWN ATOM OR ION | | 著者 | Kisiala, M, Kowalska, M, Korza, H, Czapinska, H, Bochtler, M. | | 登録日 | 2019-06-30 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Restriction endonucleases that cleave RNA/DNA heteroduplexes bind dsDNA in A-like conformation.
Nucleic Acids Res., 48, 2020
|
|
2Q7U
 
 | |
3RTV
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a closed ternary complex with natural primer/template DNA | | 分子名称: | (5'-D(*AP*AP*AP*GP*CP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DDG))-3'), 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, ... | | 著者 | Marx, A, Diederichs, K, Betz, K. | | 登録日 | 2011-05-04 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
8B4K
 
 | |
6ZY6
 
 | | Cryo-EM structure of the Human topoisomerase II alpha DNA-binding/cleavage domain in State 2 | | 分子名称: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | 著者 | Vanden Broeck, A, Lamour, V. | | 登録日 | 2020-07-30 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
6ZY5
 
 | | Cryo-EM structure of the Human topoisomerase II alpha DNA-binding/cleavage domain in State 1 | | 分子名称: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | 著者 | Vanden Broeck, A, Lamour, V. | | 登録日 | 2020-07-30 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
8C7T
 
 | |
1IXC
 
 | | Crystal structure of CbnR, a LysR family transcriptional regulator | | 分子名称: | LysR-type regulatory protein | | 著者 | Muraoka, S, Okumura, R, Ogawa, N, Miyashita, K, Senda, T. | | 登録日 | 2002-06-18 | | 公開日 | 2003-06-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of a Full-length LysR-type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend
J.Mol.Biol., 328, 2003
|
|
1IZ1
 
 | | CRYSTAL STRUCTURE OF CBNR, A LYSR FAMILY TRANSCRIPTIONAL REGULATOR | | 分子名称: | LysR-type regulatory protein | | 著者 | Muraoka, S, Okumura, R, Ogawa, N, Miyashita, K, Senda, T. | | 登録日 | 2002-09-18 | | 公開日 | 2003-05-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of a Full-length LysR-type Transcriptional Regulator, CbnR: Unusual Combination of Two Subunit Forms and Molecular Bases for Causing and Changing DNA Bend
J.Mol.Biol., 328, 2003
|
|
4XR0
 
 | |
6VGD
 
 | |
8FTJ
 
 | | Crystal structure of human NEIL1 (P2G (242K) C(delta)100) glycosylase bound to DNA duplex containing urea | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*TP*CP*CP*AP*UDV*GP*TP*CP*TP*AP*CP)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*AP*TP*GP*GP*AP*CP*GP*G)-3'), ... | | 著者 | Tomar, R, Sharma, P, Harp, J.M, Egli, M, Stone, M.P. | | 登録日 | 2023-01-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Base excision repair of the N-(2-deoxy-d-erythro-pentofuranosyl)-urea lesion by the hNEIL1 glycosylase.
Nucleic Acids Res., 51, 2023
|
|
5C8E
 
 | |
6VGG
 
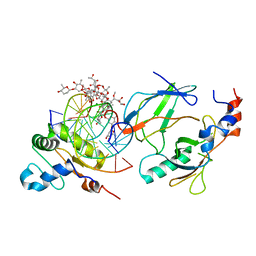 | | Crystal structure of the DNA binding domains of human transcription factor ERG, human Runx2 bound to core binding factor beta (Cbfb), and mithramycin, in complex with 16mer DNA CAGAGGATGTGGCTTC | | 分子名称: | Core-binding factor subunit beta, DNA (5'-D(P*CP*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*TP*G)-3'), ... | | 著者 | Hou, C, Rohr, J, Tsodikov, O.V. | | 登録日 | 2020-01-08 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (4.31 Å) | | 主引用文献 | Allosteric interference in oncogenic FLI1 and ERG transactions by mithramycins.
Structure, 29, 2021
|
|
8UH7
 
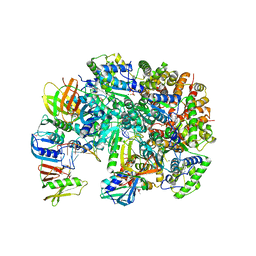 | | Structure of T4 Bacteriophage clamp loader bound to the T4 clamp, primer-template DNA, and ATP analog | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Primer DNA strand, ... | | 著者 | Gee, C.L, Marcus, K, Kelch, B.A, Makino, D.L. | | 登録日 | 2023-10-07 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.628 Å) | | 主引用文献 | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UNF
 
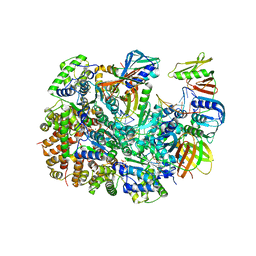 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp and DNA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Sliding clamp, ... | | 著者 | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | 登録日 | 2023-10-18 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6VGE
 
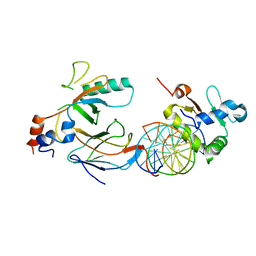 | | Crystal structure of the DNA binding domains of human transcription factor ERG, human Runx2 bound to core binding factor beta (Cbfb), in complex with 16mer DNA CAGAGGATGTGGCTTC | | 分子名称: | Core-binding factor subunit beta, DNA (5'-D(P*CP*AP*GP*AP*GP*GP*AP*TP*GP*TP*GP*GP*CP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*GP*CP*CP*AP*CP*AP*TP*CP*CP*TP*CP*TP*G)-3'), ... | | 著者 | Hou, C, Tsodikov, O.V. | | 登録日 | 2020-01-07 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (4.25 Å) | | 主引用文献 | Allosteric interference in oncogenic FLI1 and ERG transactions by mithramycins.
Structure, 29, 2021
|
|
5FDK
 
 | |
4YWM
 
 | |
4YWK
 
 | |
3SZ2
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dG as templating nucleobase | | 分子名称: | (5'-D(*AP*AP*AP*GP*CP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DDG))-3'), DNA polymerase I, ... | | 著者 | Betz, K, Marx, A, Diederichs, K. | | 登録日 | 2011-07-18 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
4YWL
 
 | |
3SV4
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dT as templating nucleobase | | 分子名称: | (5'-D(*AP*AP*AP*TP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), DNA polymerase I, ... | | 著者 | Betz, K, Diederichs, K, Marx, A. | | 登録日 | 2011-07-12 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
7X8C
 
 | | Crystal structure of a KTSC family protein from Euryarchaeon Methanolobus vulcani | | 分子名称: | KTSC domain-containing protein, SODIUM ION | | 著者 | Zhang, Z.F, Zhu, K.L, Chen, Y.Y, Cao, P, Gong, Y. | | 登録日 | 2022-03-12 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Biochemical and structural characterization of a KTSC family single-stranded DNA-binding protein from Euryarchaea.
Int.J.Biol.Macromol., 216, 2022
|
|
4NLG
 
 | | Y-family DNA polymerase chimera Dbh-Dpo4(243-245)-Dbh | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*CP*AP*CP*TP*GP*AP*TP*CP*GP*GP*G)-3', 5'-D(*TP*TP*AP*CP*GP*CP*CP*CP*TP*GP*AP*TP*CP*AP*GP*TP*GP*CP*C)-3', ... | | 著者 | Mukherjee, P, Wilson, R.C, Pata, J.D. | | 登録日 | 2013-11-14 | | 公開日 | 2014-01-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Three Residues of the Interdomain Linker Determine the Conformation and Single-base Deletion Fidelity of Y-family Translesion Polymerases.
J.Biol.Chem., 289, 2014
|
|
