1W34
 
 | | FERREDOXIN-NADP REDUCTASE (MUTATION: Y 303 S) | | 分子名称: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | 著者 | Hermoso, J.A, Perez-Dorado, I, Medina, M, Julvez, M.M, Sanz-Aparicio, J, Gomez-Moreno, C. | | 登録日 | 2004-07-13 | | 公開日 | 2005-10-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | C-Terminal Tyrosine of Ferredoxin-Nadp(+) Reductase in Hydride Transfer Processes with Nad(P)(+)/H.
Biochemistry, 44, 2005
|
|
1SN6
 
 | |
3BUM
 
 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in Sprouty2 | | 分子名称: | E3 ubiquitin-protein ligase CBL, Protein sprouty homolog 2 | | 著者 | Ng, C, Jackson, A.R, Buschdorf, P.J, Sun, Q, Guy, R.G, Sivaraman, J. | | 登録日 | 2008-01-03 | | 公開日 | 2008-02-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
4BSM
 
 | | Crystal structure of the Nuclear Export Receptor CRM1 (exportin-1) lacking the C-terminal helical extension at 4.5A | | 分子名称: | EXPORTIN-1 | | 著者 | Dian, C, Bernaudat, F, Langer, K, Oliva, M.F, Fornerod, M, Schoehn, G, Muller, C.W, Petosa, C. | | 登録日 | 2013-06-10 | | 公開日 | 2013-07-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (4.5 Å) | | 主引用文献 | Structure of a Truncation Mutant of the Nuclear Export Factor Crm1 Provides Insights Into the Auto-Inhibitory Role of its C-Terminal Helix.
Structure, 21, 2013
|
|
4BSN
 
 | | Crystal structure of the Nuclear Export Receptor CRM1 (exportin-1) lacking the C-terminal helical extension at 4.1A | | 分子名称: | EXPORTIN-1 | | 著者 | Dian, C, Bernaudat, F, Langer, K, Oliva, M.F, Fornerod, M, Schoehn, G, Muller, C.W, Petosa, C. | | 登録日 | 2013-06-11 | | 公開日 | 2013-07-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (4.1 Å) | | 主引用文献 | Structure of a Truncation Mutant of the Nuclear Export Factor Crm1 Provides Insights Into the Auto-Inhibitory Role of its C-Terminal Helix.
Structure, 21, 2013
|
|
3VKE
 
 | |
2G15
 
 | | Structural Characterization of autoinhibited c-Met kinase | | 分子名称: | activated met oncogene | | 著者 | Wang, W, Marimuthu, A, Tsai, J, Kumar, A, Krupka, H.I, Zhang, C, Powell, B, Suzuki, Y, Nguyen, H, Tabrizizad, M, Luu, C, West, B.L. | | 登録日 | 2006-02-13 | | 公開日 | 2006-03-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural characterization of autoinhibited c-Met kinase produced by coexpression in bacteria with phosphatase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2G01
 
 | | Pyrazoloquinolones as Novel, Selective JNK1 inhibitors | | 分子名称: | 6-CHLORO-9-HYDROXY-1,3-DIMETHYL-1,9-DIHYDRO-4H-PYRAZOLO[3,4-B]QUINOLIN-4-ONE, C-jun-amino-terminal kinase-interacting protein 1, Mitogen-activated protein kinase 8, ... | | 著者 | Abad-Zapatero, C. | | 登録日 | 2006-02-10 | | 公開日 | 2006-04-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Synthesis and SAR of 1,9-dihydro-9-hydroxypyrazolo[3,4-b]quinolin-4-ones as novel, selective c-Jun N-terminal kinase inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1UX6
 
 | |
3CCN
 
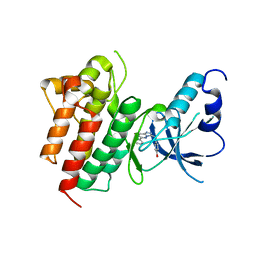 | | X-ray structure of c-Met with triazolopyridazine inhibitor. | | 分子名称: | 4-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methyl]phenol, Hepatocyte growth factor receptor | | 著者 | Abrecht, B.K, Harmange, J.-C, Bauer, D, Dussault, I, long, A, Bellon, S.F. | | 登録日 | 2008-02-26 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery and Optimization of Triazolopyridazines as Potent and Selective Inhibitors of the c-Met Kinase.
J.Med.Chem., 51, 2008
|
|
3RHK
 
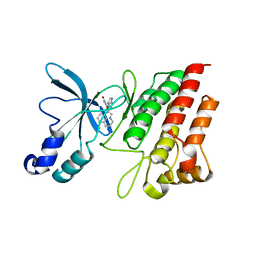 | | Crystal structure of the catalytic domain of c-Met kinase in complex with ARQ 197 | | 分子名称: | 1-[(3R,4R)-4-(1H-indol-3-yl)-2,5-dioxopyrrolidin-3-yl]pyrrolo[3,2,1-ij]quinolinium, Hepatocyte growth factor receptor | | 著者 | Eathiraj, S, Palma, R, Volckova, E, Hirschi, M, France, D.S, Ashwell, M.A, Chan, T.C. | | 登録日 | 2011-04-11 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Discovery of a novel mode of protein kinase inhibition characterized by the mechanism of inhibition of human mesenchymal-epithelial transition factor (c-Met) protein autophosphorylation by ARQ 197.
J.Biol.Chem., 286, 2011
|
|
3CD8
 
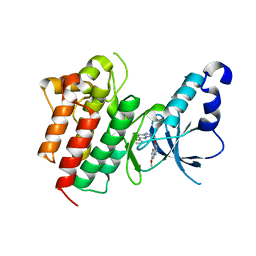 | | X-ray Structure of c-Met with triazolopyridazine Inhibitor. | | 分子名称: | 7-methoxy-4-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methoxy]quinoline, Hepatocyte growth factor receptor | | 著者 | Bellon, S.F, Albrecht, B.K, Harmange, J.-C, Bauer, D, Choquette, D, Dussault, I. | | 登録日 | 2008-02-26 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery and Optimization of Triazolopyridazines as Potent and Selective Inhibitors of the c-Met Kinase.
J.Med.Chem., 51, 2008
|
|
1TDB
 
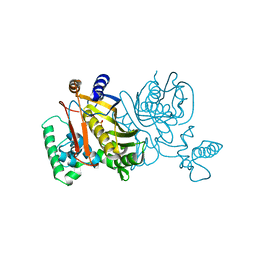 | | STRUCTURES OF THYMIDYLATE SYNTHASE WITH A C-TERMINAL DELETION: ROLE OF THE C-TERMINUS IN ALIGNMENT OF D/UMP AND CH2H4FOLATE | | 分子名称: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | 著者 | Perry, K.M, Carreras, C.W, Chang, L.C, Santi, D.V, Stroud, R.M. | | 登録日 | 1993-02-15 | | 公開日 | 1993-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structures of thymidylate synthase with a C-terminal deletion: role of the C-terminus in alignment of 2'-deoxyuridine 5'-monophosphate and 5,10-methylenetetrahydrofolate.
Biochemistry, 32, 1993
|
|
1T0C
 
 | |
1KNJ
 
 | | Co-Crystal Structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate Synthase (ispF) from E. coli Involved in Mevalonate-Independent Isoprenoid Biosynthesis, Complexed with CMP/MECDP/Mn2+ | | 分子名称: | 2C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE, 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-MONOPHOSPHATE, ... | | 著者 | Richard, S.B, Ferrer, J.L, Bowman, M.E, Lillo, A.M, Tetzlaff, C.N, Cane, D.E, Noel, J.P. | | 登録日 | 2001-12-18 | | 公開日 | 2002-06-18 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure and mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase. An enzyme in the mevalonate-independent isoprenoid biosynthetic pathway.
J.Biol.Chem., 277, 2002
|
|
1TDA
 
 | | STRUCTURES OF THYMIDYLATE SYNTHASE WITH A C-TERMINAL DELETION: ROLE OF THE C-TERMINUS IN ALIGNMENT OF D/UMP AND CH2H4FOLATE | | 分子名称: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | 著者 | Perry, K.M, Carreras, C.W, Chang, L.C, Santi, D.V, Stroud, R.M. | | 登録日 | 1993-02-15 | | 公開日 | 1993-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Structures of thymidylate synthase with a C-terminal deletion: role of the C-terminus in alignment of 2'-deoxyuridine 5'-monophosphate and 5,10-methylenetetrahydrofolate.
Biochemistry, 32, 1993
|
|
3ZJZ
 
 | | Open-form NavMS Sodium Channel Pore (with C-terminal Domain) | | 分子名称: | DODECAETHYLENE GLYCOL, HEGA-10, ION TRANSPORT PROTEIN, ... | | 著者 | Bagneris, C, Naylor, C.E, Wallace, B.A. | | 登録日 | 2013-01-21 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | Role of the C-Terminal Domain in the Structure and Function of Tetrameric Sodium Channels.
Nat.Commun., 4, 2013
|
|
1MUZ
 
 | |
1EGI
 
 | | STRUCTURE OF A C-TYPE CARBOHYDRATE-RECOGNITION DOMAIN (CRD-4) FROM THE MACROPHAGE MANNOSE RECEPTOR | | 分子名称: | CALCIUM ION, MACROPHAGE MANNOSE RECEPTOR | | 著者 | Feinberg, H, Park-Snyder, S, Kolatkar, A.R, Heise, C.T, Taylor, M.E, Weis, W.I. | | 登録日 | 2000-02-15 | | 公開日 | 2000-08-30 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of a C-type carbohydrate recognition domain from the macrophage mannose receptor.
J.Biol.Chem., 275, 2000
|
|
1MV0
 
 | |
2AVF
 
 | | Crystal Structure of C-terminal Desundecapeptide Nitrite Reductase from Achromobacter cycloclastes | | 分子名称: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase | | 著者 | Li, H.T, Chang, T, Chang, W.C, Chen, C.J, Liu, M.Y, Gui, L.L, Zhang, J.P, An, X.M, Chang, W.R. | | 登録日 | 2005-08-30 | | 公開日 | 2005-12-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of C-terminal desundecapeptide nitrite reductase from Achromobacter cycloclastes
Biochem.Biophys.Res.Commun., 338, 2005
|
|
3FRY
 
 | | Crystal structure of the CopA C-terminal metal binding domain | | 分子名称: | CITRIC ACID, Probable copper-exporting P-type ATPase A | | 著者 | Agarwal, S, Sazinsky, M, Arguello, J, Rosenzweig, A.C. | | 登録日 | 2009-01-08 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and interactions of the C-terminal metal binding domain of Archaeoglobus fulgidus CopA.
Proteins, 78, 2010
|
|
3O8I
 
 | | Structure of 14-3-3 isoform sigma in complex with a C-Raf1 peptide and a stabilizing small molecule fragment | | 分子名称: | 14-3-3 binding site peptide of RAF proto-oncogene serine/threonine-protein kinase, 14-3-3 protein sigma, 6,6-dihydroxy-1-methoxyhexan-2-one | | 著者 | Ottmann, C, Rose, R, Kaiser, M, Kuhenne, P. | | 登録日 | 2010-08-03 | | 公開日 | 2010-09-29 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Impaired Binding of 14-3-3 to C-RAF in Noonan Syndrome Suggests New Approaches in Diseases with Increased Ras Signaling
Mol.Cell.Biol., 30, 2010
|
|
2OCC
 
 | |
1LBK
 
 | | Crystal structure of a recombinant glutathione transferase, created by replacing the last seven residues of each subunit of the human class pi isoenzyme with the additional C-terminal helix of human class alpha isoenzyme | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, Glutathione S-transferase class pi chimaera (CODA), ... | | 著者 | Kong, G.K.W, Micaloni, C, Mazzetti, A.P, Nuccetelli, M, Antonini, G, Stella, L, McKinstry, W.J, Polekhina, G, Rossjohn, J, Federici, G, Ricci, G, Parker, M.W, Lo Bello, M. | | 登録日 | 2002-04-04 | | 公開日 | 2002-04-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Engineering a new C-terminal tail in the H-site of human glutathione transferase P1-1: structural and functional consequences.
J.Mol.Biol., 325, 2003
|
|
