5EBH
 
 | |
6UXW
 
 | | SWI/SNF nucleosome complex with ADP-BeFx | | 分子名称: | 601 sequence bottom strand, 601 sequence top strand, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | He, Y, Han, Y. | | 登録日 | 2019-11-08 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (8.96 Å) | | 主引用文献 | Cryo-EM structure of SWI/SNF complex bound to a nucleosome.
Nature, 579, 2020
|
|
4RSK
 
 | |
8CB1
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with N-PNT-DNM 15 | | 分子名称: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[5-(phenanthren-9-ylmethoxy)pentyl]piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Sulzenbacher, G, Roig-Zamboni, V, Overkleeft, H, Artola, M. | | 登録日 | 2023-01-25 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Fluorescence polarisation activity-based protein profiling for the identification of deoxynojirimycin-type inhibitors selective for lysosomal retaining alpha- and beta-glucosidases.
Chem Sci, 14, 2023
|
|
8CB6
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in covalent complex with TAMRA tagged 1,6-Epi-cylcophellitol aziridine activity based probe | | 分子名称: | (1S,2R,3R,4R,5R)-5-[8-[4-(4-azanylbutyl)-1,2,3-triazol-1-yl]octylamino]-4-(hydroxymethyl)cyclohexane-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Sulzenbacher, G, Roig-Zamboni, V, Overkleeft, H, Artola, M. | | 登録日 | 2023-01-25 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Fluorescence polarisation activity-based protein profiling for the identification of deoxynojirimycin-type inhibitors selective for lysosomal retaining alpha- and beta-glucosidases.
Chem Sci, 14, 2023
|
|
4S0G
 
 | |
5FXK
 
 | | GluN1b-GluN2B NMDA receptor structure-Class Y | | 分子名称: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | 著者 | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | 登録日 | 2016-03-02 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
1GHL
 
 | |
4TNU
 
 | |
4RNT
 
 | |
5FXG
 
 | | GLUN1B-GLUN2B NMDA RECEPTOR IN ACTIVE CONFORMATION | | 分子名称: | N-METHYL-D-ASPARTATE RECEPTOR GLUN1, N-METHYL-D-ASPARTATE RECEPTOR GLUN2B | | 著者 | Tajima, N, Karakas, E, Grant, T, Simorowski, N, Diaz-Avalos, R, Grigorieff, N, Furukawa, H. | | 登録日 | 2016-03-02 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.8 Å) | | 主引用文献 | Activation of Nmda Receptors and the Mechanism of Inhibition by Ifenprodil.
Nature, 534, 2016
|
|
5B0Z
 
 | | The crystal structure of the nucleosome containing H3.2, at 1.98 A resolution | | 分子名称: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | 著者 | Suzuki, Y, Horikoshi, N, Kato, D, Kurumizaka, H. | | 登録日 | 2015-11-14 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.987 Å) | | 主引用文献 | Crystal structure of the nucleosome containing histone H3 with crotonylated lysine 122
Biochem.Biophys.Res.Commun., 469, 2016
|
|
5HK5
 
 | |
8C84
 
 | | Crystal structure of MADS-box/MEF2D N-terminal domain complex | | 分子名称: | DNA (5'-D(P*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*TP*T)-3'), MEF2D protein | | 著者 | Chinellato, M, Carli, A, Perin, S, Mazzoccato, Y, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | 登録日 | 2023-01-18 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
6UXV
 
 | | SWI/SNF Body Module | | 分子名称: | SWI/SNF chromatin-remodeling complex subunit SNF5, SWI/SNF chromatin-remodeling complex subunit SWI1, SWI/SNF complex subunit SWI3, ... | | 著者 | He, Y, Han, Y. | | 登録日 | 2019-11-08 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Cryo-EM structure of SWI/SNF complex bound to a nucleosome.
Nature, 579, 2020
|
|
1RNZ
 
 | | RIBONUCLEASE A CRYSTALLIZED FROM 2.5M SODIUM CHLORIDE, 3.3M SODIUM FORMATE | | 分子名称: | CHLORIDE ION, RIBONUCLEASE A | | 著者 | Fedorov, A.A, Josph-Mccarthy, D, Fedorov, E.V, Sirakova, D, Graf, I, Almo, S.C. | | 登録日 | 1996-11-08 | | 公開日 | 1997-04-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Ionic interactions in crystalline bovine pancreatic ribonuclease A.
Biochemistry, 35, 1996
|
|
5B0Y
 
 | | Crystal structure of the nucleosome containing histone H3 with the crotonylated lysine 122 | | 分子名称: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | 著者 | Suzuki, Y, Horikoshi, N, Kurumizaka, H. | | 登録日 | 2015-11-13 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.557 Å) | | 主引用文献 | Crystal structure of the nucleosome containing histone H3 with crotonylated lysine 122
Biochem.Biophys.Res.Commun., 469, 2016
|
|
8C58
 
 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 5-hydroxycytosine and 5-methylcytosine containing dsDNA | | 分子名称: | CARBONATE ION, Cytosine-specific methyltransferase, DNA (5'-D(*CP*CP*AP*CP*AP*TP*GP*(5OC)P*GP*CP*TP*GP*AP*A)-3'), ... | | 著者 | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | 登録日 | 2023-01-06 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
8C59
 
 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 5-bromocytosine (converted to 5mC) and 5-methylcytosine containing dsDNA | | 分子名称: | CARBONATE ION, CITRIC ACID, Cytosine-specific methyltransferase, ... | | 著者 | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | 登録日 | 2023-01-06 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
8C57
 
 | | CpG specific M.MpeI methyltransferase crystallized in the presence of 5,6-dihydro-5-azacytosine (converted to 5m-dhaC) and 5-methylcytosine containing dsDNA | | 分子名称: | CARBONATE ION, Cytosine-specific methyltransferase, DNA (5'-D(*CP*CP*AP*CP*AP*TP*GP*(5MA)P*GP*CP*TP*GP*AP*A)-3'), ... | | 著者 | Wojciechowski, M, Czapinska, H, Krwawicz, J, Rafalski, D, Bochtler, M. | | 登録日 | 2023-01-06 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Cytosine analogues as DNA methyltransferase substrates.
Nucleic Acids Res., 52, 2024
|
|
3HT8
 
 | | 5-chloro-2-methylphenol in complex with T4 lysozyme L99A/M102Q | | 分子名称: | 5-chloro-2-methylphenol, Lysozyme, PHOSPHATE ION | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-11 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HTG
 
 | | 2-ethoxy-3,4-dihydro-2h-pyran in complex with T4 lysozyme L99A/M102Q | | 分子名称: | (2S)-2-ethoxy-3,4-dihydro-2H-pyran, BETA-MERCAPTOETHANOL, Lysozyme, ... | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-11 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HUA
 
 | | 4,5,6,7-tetrahydroindole in complex with T4 lysozyme L99A/M102Q | | 分子名称: | 4,5,6,7-tetrahydro-1H-indole, BETA-MERCAPTOETHANOL, Lysozyme, ... | | 著者 | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | 登録日 | 2009-06-13 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
3HKR
 
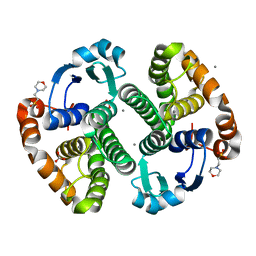 | | Crystal Structure of Glutathione Transferase Pi Y108V Mutant | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CARBONATE ION, ... | | 著者 | Parker, L.J. | | 登録日 | 2009-05-25 | | 公開日 | 2009-09-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Influence of the H-site residue 108 on human glutathione transferase P1-1 ligand binding: structure-thermodynamic relationships and thermal stability.
Protein Sci., 18, 2009
|
|
6TJM
 
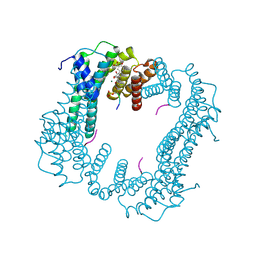 | | Crystal structure of an Estrogen Receptor alpha 8-mer phosphopeptide in complex with 14-3-3sigma stabilized by Pyrrolidone1 | | 分子名称: | 14-3-3 protein sigma, 5-[(2~{R})-2-(4-nitrophenyl)-4-oxidanyl-5-oxidanylidene-3-(phenylcarbonyl)-2~{H}-pyrrol-1-yl]-2-oxidanyl-benzoic acid, C-terminal phosphopeptide of human estrogen receptor alpha, ... | | 著者 | Andrei, S.A, Bosica, F, Ottmann, C, O'Mahony, G. | | 登録日 | 2019-11-26 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Design of Drug-Like Protein-Protein Interaction Stabilizers Guided By Chelation-Controlled Bioactive Conformation Stabilization.
Chemistry, 26, 2020
|
|
