8PFI
 
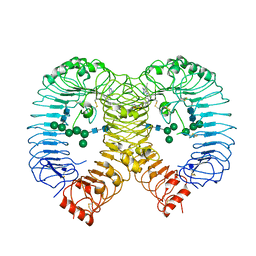 | | Crystal structure of human TLR8 in complex with compound 34 | | 分子名称: | (3~{S})-~{N}-[4-[[5-(1,6-dimethylpyrazolo[3,4-b]pyridin-4-yl)-3-methyl-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-1-yl]methyl]-1-bicyclo[2.2.2]octanyl]morpholine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | 著者 | Faller, M, Zink, F. | | 登録日 | 2023-06-16 | | 公開日 | 2023-08-16 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.785 Å) | | 主引用文献 | Discovery of the TLR7/8 Antagonist MHV370 for Treatment of Systemic Autoimmune Diseases.
Acs Med.Chem.Lett., 14, 2023
|
|
8F77
 
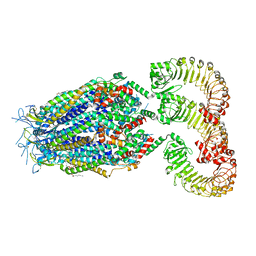 | | LRRC8A(T48D):C conformation 2 top focus | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | 著者 | Kern, D.M, Brohawn, S.G. | | 登録日 | 2022-11-18 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-07-05 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4WLP
 
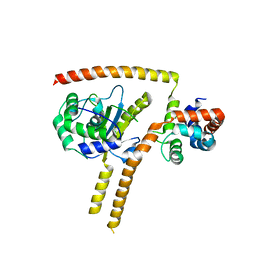 | | Crystal structure of UCH37-NFRKB Inhibited Deubiquitylating Complex | | 分子名称: | Nuclear factor related to kappa-B-binding protein, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | 著者 | Hemmis, C.W, Hill, C.P, VanderLinden, R, Whitby, F.G. | | 登録日 | 2014-10-07 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.102 Å) | | 主引用文献 | Structural Basis for the Activation and Inhibition of the UCH37 Deubiquitylase.
Mol.Cell, 57, 2015
|
|
6N3O
 
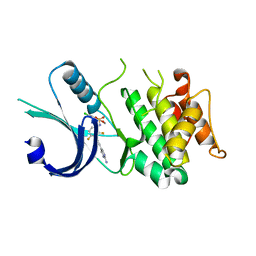 | | Identification of novel, potent and selective GCN2 inhibitors as first-in-class anti-tumor agents | | 分子名称: | N-{3-[(2-aminopyrimidin-5-yl)ethynyl]-2,4-difluorophenyl}-5-chloro-2-methoxypyridine-3-sulfonamide, eIF-2-alpha kinase GCN2 | | 著者 | Hoffman, I.D, Fujimoto, J, Kurasawa, O, Takagi, T, Klein, M.G, Kefala, G, Ding, S.C, Cary, D.R, Mizojiri, R. | | 登録日 | 2018-11-15 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Identification of Novel, Potent, and Orally Available GCN2 Inhibitors with Type I Half Binding Mode.
Acs Med.Chem.Lett., 10, 2019
|
|
7ZO8
 
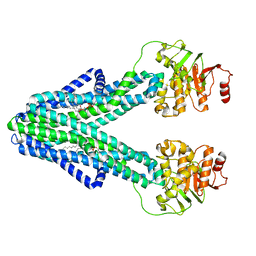 | | cryo-EM structure of CGT ABC transporter in nanodisc apo state | | 分子名称: | Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, DIUNDECYL PHOSPHATIDYL CHOLINE | | 著者 | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | 登録日 | 2022-04-24 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5N4C
 
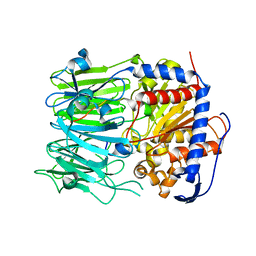 | | Prolyl oligopeptidase B from Galerina marginata bound to 35mer hydrolysis and macrocyclization substrate - S577A mutant | | 分子名称: | Alpha-amanitin proprotein, GLYCEROL, Prolyl oligopeptidase | | 著者 | Czekster, C.M, McMahon, S.A, Ludewig, H, Naismith, J.H. | | 登録日 | 2017-02-10 | | 公開日 | 2017-11-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates.
Nat Commun, 8, 2017
|
|
7ZO9
 
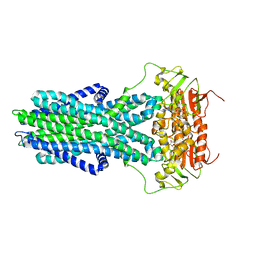 | | cryo-EM structure of CGT ABC transporter in vanadate trapped state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, VANADATE ION | | 著者 | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | 登録日 | 2022-04-24 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZNU
 
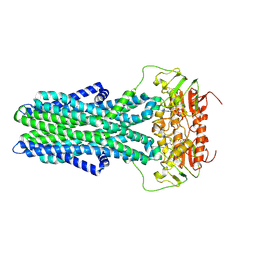 | | cryo-EM structure of CGT ABC transporter in detergent micelle | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, VANADATE ION | | 著者 | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | 登録日 | 2022-04-22 | | 公開日 | 2022-12-07 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8I4Z
 
 | | CalA3 with hydrolysis product | | 分子名称: | 11-oxidanylidene-11-(1~{H}-pyrrol-2-yl)undecanoic acid, Beta-ketoacyl-acyl-carrier-protein synthase I | | 著者 | Wang, J, Wang, Z. | | 登録日 | 2023-01-21 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.97 Å) | | 主引用文献 | C-N bond formation by a polyketide synthase.
Nat Commun, 14, 2023
|
|
7UGA
 
 | | Solution structure of NPSL2 | | 分子名称: | NPSL2 RNA (43-MER) | | 著者 | Liu, Y, Keane, S.C. | | 登録日 | 2022-03-24 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Solution Structure of NPSL2, A Regulatory Element in the oncomiR-1 RNA.
J.Mol.Biol., 434, 2022
|
|
8R9T
 
 | |
6SO9
 
 | |
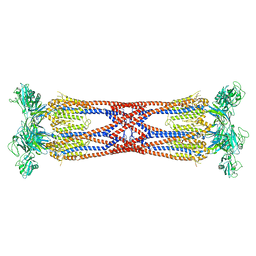
6SIH
 
 | |
7N0B
 
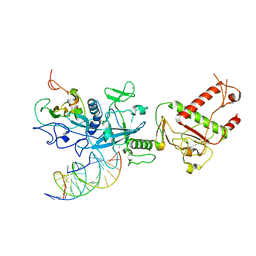 | |
8OKM
 
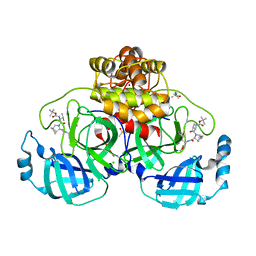 | | Crystal structure of F2F-2020197-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-03-28 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
6H0K
 
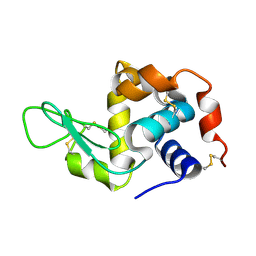 | | Hen egg-white lysozyme structure determined with data from the EuXFEL, the first MHz free electron laser, 7.47 keV photon energy | | 分子名称: | Lysozyme C | | 著者 | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | 登録日 | 2018-07-10 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6H0L
 
 | | Hen egg-white lysozyme structure determined with data from the EuXFEL, 9.22 keV photon energy | | 分子名称: | Lysozyme C | | 著者 | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | 登録日 | 2018-07-10 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6OHW
 
 | | Structural basis for human coronavirus attachment to sialic acid receptors. Apo-HCoV-OC43 S | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike surface glycoprotein, ... | | 著者 | Tortorici, M.A, Walls, A.C, Lang, Y, Wang, C, Li, Z, Koerhuis, D, Boons, G.J, Bosch, B.J, Rey, F.A, de Groot, R, Veesler, D. | | 登録日 | 2019-04-07 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for human coronavirus attachment to sialic acid receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4RDZ
 
 | | Crystal structure of VmoLac in P64 space group | | 分子名称: | COBALT (II) ION, GLYCEROL, MYRISTIC ACID, ... | | 著者 | Hiblot, J, Bzdrenga, J, Champion, C, Gotthard, G, Gonzalez, D, Chabriere, E, Elias, M. | | 登録日 | 2014-09-20 | | 公開日 | 2015-02-25 | | 最終更新日 | 2025-03-26 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of VmoLac, a tentative quorum quenching lactonase from the extremophilic crenarchaeon Vulcanisaeta moutnovskia.
Sci Rep, 5, 2015
|
|
8TGX
 
 | | Crystal structure of C. elegans LGG-1 | | 分子名称: | CHLORIDE ION, Protein lgg-1 | | 著者 | Cheung, Y.W.S, Yip, C.K. | | 登録日 | 2023-07-13 | | 公開日 | 2025-01-22 | | 最終更新日 | 2025-05-28 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Structure of the human autophagy factor EPG5 and the molecular basis of its conserved mode of interaction with Atg8-family proteins.
Autophagy, 21, 2025
|
|
1UNL
 
 | | Structural mechanism for the inhibition of CD5-p25 from the roscovitine, aloisine and indirubin. | | 分子名称: | CYCLIN-DEPENDENT KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1, R-ROSCOVITINE | | 著者 | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | 登録日 | 2003-09-10 | | 公開日 | 2004-11-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
1URO
 
 | | UROPORPHYRINOGEN DECARBOXYLASE | | 分子名称: | BETA-MERCAPTOETHANOL, PROTEIN (UROPORPHYRINOGEN DECARBOXYLASE) | | 著者 | Whitby, F.G, Phillips, J.D, Kushner, J.P, Hill, C.P. | | 登録日 | 1998-08-21 | | 公開日 | 1998-08-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of human uroporphyrinogen decarboxylase.
EMBO J., 17, 1998
|
|
8K8M
 
 | |
8ZRH
 
 | | HBcAg-D4 Fab complex | | 分子名称: | Capsid protein, Heavy chains of D4 Fab, Light chains of D4 Fab | | 著者 | Zhang, Z, Ju, B, Liu, C, Yan, H. | | 登録日 | 2024-06-04 | | 公開日 | 2024-11-13 | | 最終更新日 | 2025-02-19 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The characterization and structural basis of a human broadly binding antibody to HBV core protein.
J.Virol., 99, 2025
|
|
6O47
 
 | | human cGAS core domain (K427E/K428E) bound with RU-521 | | 分子名称: | (3~{S})-3-[1-[4,5-bis(chloranyl)-1~{H}-benzimidazol-2-yl]-3-methyl-5-oxidanyl-pyrazol-4-yl]-3~{H}-2-benzofuran-1-one, 2-(4,5-dichloro-1H-benzimidazol-2-yl)-5-methyl-4-[(1R)-3-oxo-1,3-dihydro-2-benzofuran-1-yl]-1,2-dihydro-3H-pyrazol-3-one, CITRIC ACID, ... | | 著者 | Xie, W, Lama, L, Adura, C, Glickman, J.F, Tuschl, T, Patel, D.J. | | 登録日 | 2019-02-28 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.196 Å) | | 主引用文献 | Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
