9BKD
 
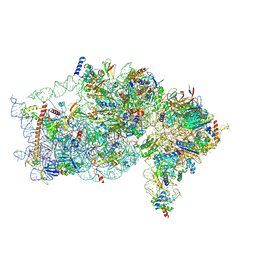 | | The structure of human Pdcd4 bound to the 40S small ribosomal subunit | | 分子名称: | 18S rRNA, 40S ribosomal protein S12, 40S ribosomal protein S13, ... | | 著者 | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Zuber, P, Albacete-Albacete, L, Ramakrishnan, V, S.Fraser, C. | | 登録日 | 2024-04-27 | | 公開日 | 2024-09-04 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Human tumor suppressor protein Pdcd4 binds at the mRNA entry channel in the 40S small ribosomal subunit.
Nat Commun, 15, 2024
|
|
6TMO
 
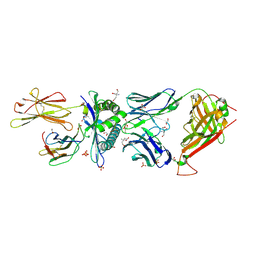 | | Structure determination of an enhanced affinity TCR, a24b17, in complex with HLA-A*02:01 presenting a MART-1 peptide, EAAGIGILTV | | 分子名称: | 1,2-ETHANEDIOL, Alpha chain of engineered high affinity T-cell receptor, Beta chain of high affinity engineered T-cell receptor, ... | | 著者 | Rizkallah, P.J, Cole, D.K. | | 登録日 | 2019-12-05 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Molecular Rules Underpinning Enhanced Affinity Binding of Human T Cell Receptors Engineered for Immunotherapy.
Mol Ther Oncolytics, 18, 2020
|
|
9BLN
 
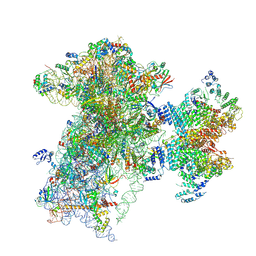 | | The structure of human Pdcd4 bound to the 40S-eIF4A-eIF3-eIF1 complex | | 分子名称: | 40S ribosomal protein S14, ACETIC ACID, Eukaryotic initiation factor 4A-I, ... | | 著者 | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Zuber, P, Yifei, D, Albacete-Albacete, L, Ramakrishnan, V, S Fraser, C. | | 登録日 | 2024-04-30 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Human tumor suppressor protein Pdcd4 binds at the mRNA entry channel in the 40S small ribosomal subunit.
Nat Commun, 15, 2024
|
|
6TQO
 
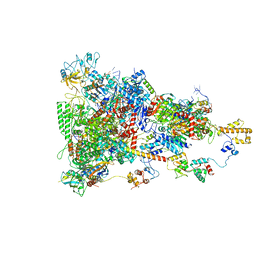 | | rrn anti-termination complex | | 分子名称: | 30S ribosomal protein S10, 30S ribosomal protein S4, DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Huang, Y.H, Wahl, M.C, Loll, B, Hilal, T, Said, N. | | 登録日 | 2019-12-17 | | 公開日 | 2020-08-05 | | 最終更新日 | 2025-07-09 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis.
Mol.Cell, 79, 2020
|
|
6TR9
 
 | | Crystal structure of human L ferritin (HuLf) triple variant E60A-E61A-E64A | | 分子名称: | CADMIUM ION, Ferritin light chain | | 著者 | Pozzi, C, Ciambellotti, S, Turano, P, Mangani, S. | | 登録日 | 2019-12-18 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Iron Biomineral Growth from the Initial Nucleation Seed in L-Ferritin.
Chemistry, 26, 2020
|
|
6TS1
 
 | | Crystal structure of human L ferritin (HuLf) triple variant E60A-E61A-E64A Fe(III)-loaded for 60 minutes | | 分子名称: | CADMIUM ION, FE (III) ION, Ferritin light chain, ... | | 著者 | Pozzi, C, Ciambellotti, S, Turano, P, Mangani, S. | | 登録日 | 2019-12-19 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Iron Biomineral Growth from the Initial Nucleation Seed in L-Ferritin.
Chemistry, 26, 2020
|
|
6TSJ
 
 | | Crystal structure of human L ferritin (HuLf) Fe(III)-loaded for 15 minutes | | 分子名称: | CADMIUM ION, FE (III) ION, Ferritin light chain, ... | | 著者 | Pozzi, C, Ciambellotti, S, Turano, P, Mangani, S. | | 登録日 | 2019-12-20 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Iron Biomineral Growth from the Initial Nucleation Seed in L-Ferritin.
Chemistry, 26, 2020
|
|
6TM1
 
 | |
6TRO
 
 | | Crystal structure of the T-cell receptor GVY01 bound to HLA A2*01-GVYDGREHTV | | 分子名称: | Beta-2-microglobulin, MAGE-A4 peptide (amino acids 230-239), MHC class I antigen, ... | | 著者 | Coles, C.H, McMurran, C, Lloyd, A, Hibbert, L, Lupardus, P.J, Cole, D.K, Harper, S. | | 登録日 | 2019-12-19 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | T cell receptor interactions with human leukocyte antigen govern indirect peptide selectivity for the cancer testis antigen MAGE-A4.
J.Biol.Chem., 295, 2020
|
|
6TTP
 
 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 1/Adenosine (DHU_M3M_023) | | 分子名称: | ACETATE ION, ADENOSINE, N6-adenosine-methyltransferase catalytic subunit, ... | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-12-30 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
6TRN
 
 | | Crystal structure of HLA A2*01-AVYDGREHTV | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, MAGE-A4 peptide (amino acids 230-239) variant, ... | | 著者 | Coles, C.H, McMurran, C, Lloyd, A, Hibbert, L, Lupardus, P.J, Cole, D.K, Harper, S. | | 登録日 | 2019-12-19 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | T cell receptor interactions with human leukocyte antigen govern indirect peptide selectivity for the cancer testis antigen MAGE-A4.
J.Biol.Chem., 295, 2020
|
|
6TSF
 
 | | Crystal structure of human L ferritin (HuLf) Fe(III)-loaded for 60 minutes | | 分子名称: | CADMIUM ION, FE (III) ION, Ferritin light chain, ... | | 著者 | Pozzi, C, Ciambellotti, S, Turano, P, Mangani, S. | | 登録日 | 2019-12-20 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Iron Biomineral Growth from the Initial Nucleation Seed in L-Ferritin.
Chemistry, 26, 2020
|
|
6TSA
 
 | | Crystal structure of human L ferritin (HuLf) Fe(III)-loaded for 30 minutes | | 分子名称: | CADMIUM ION, FE (III) ION, Ferritin light chain, ... | | 著者 | Pozzi, C, Ciambellotti, S, Turano, P, Mangani, S. | | 登録日 | 2019-12-20 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Iron Biomineral Growth from the Initial Nucleation Seed in L-Ferritin.
Chemistry, 26, 2020
|
|
6N7X
 
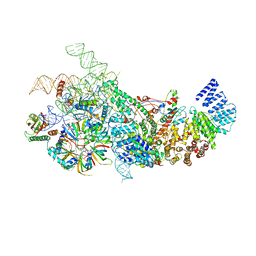 | | S. cerevisiae U1 snRNP | | 分子名称: | 56 kDa U1 small nuclear ribonucleoprotein component, Pre-mRNA-processing factor 39, Protein NAM8, ... | | 著者 | Li, X, Liu, S, Jiang, J, Zhang, L, Espinosa, S, Hill, R.C, Hansen, K.C, Zhou, Z.H, Zhao, R. | | 登録日 | 2018-11-28 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | CryoEM structure of Saccharomyces cerevisiae U1 snRNP offers insight into alternative splicing.
Nat Commun, 8, 2017
|
|
1H3L
 
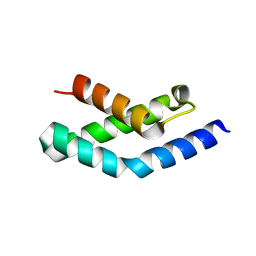 | | N-terminal fragment of SigR from Streptomyces coelicolor | | 分子名称: | RNA POLYMERASE SIGMA FACTOR | | 著者 | Li, W, Stevenson, C.E.M, Burton, N, Jakimowicz, P, Paget, M.S.B, Buttner, M.J, Lawson, D.M, Kleanthous, C. | | 登録日 | 2002-09-10 | | 公開日 | 2002-10-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.375 Å) | | 主引用文献 | Identification and Structure of the Anti-Sigma Factor-Binding Domain of the Disulfide-Stress Regulated Sigma Factor Sigma(R) from Streptomyces Coelicolor
J.Mol.Biol., 323, 2002
|
|
6ZM7
 
 | | SARS-CoV-2 Nsp1 bound to the human CCDC124-80S-EBP1 ribosome complex | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-01 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZMO
 
 | | SARS-CoV-2 Nsp1 bound to the human LYAR-80S-eEF1a ribosome complex | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-03 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZMI
 
 | | SARS-CoV-2 Nsp1 bound to the human LYAR-80S ribosome complex | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-02 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
7JLU
 
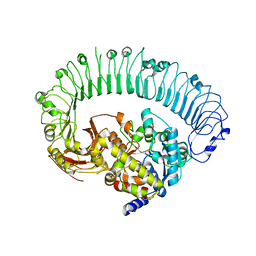 | | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ | | 分子名称: | CALCIUM ION, Disease resistance protein Roq1, XopQ | | 著者 | Martin, R, Qi, T, Zhang, H, Lui, F, King, M, Toth, C, Nogales, E, Staskawicz, B.J. | | 登録日 | 2020-07-30 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ.
Science, 370, 2020
|
|
7MQ8
 
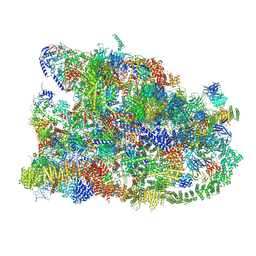 | | Cryo-EM structure of the human SSU processome, state pre-A1 | | 分子名称: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | 著者 | Vanden Broeck, A, Singh, S, Klinge, S. | | 登録日 | 2021-05-05 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-08-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
7MQ9
 
 | | Cryo-EM structure of the human SSU processome, state pre-A1* | | 分子名称: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | 著者 | Vanden Broeck, A, Singh, S, Klinge, S. | | 登録日 | 2021-05-05 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
4LC5
 
 | | Structural basis of substrate specificity of CDA superfamily guanine deaminase | | 分子名称: | 1,2-ETHANEDIOL, 9-METHYLGUANINE, Cytidine and deoxycytidylate deaminase zinc-binding region, ... | | 著者 | Bitra, A, Biswas, A, Anand, R. | | 登録日 | 2013-06-21 | | 公開日 | 2014-01-22 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural basis of the substrate specificity of cytidine deaminase superfamily Guanine deaminase
Biochemistry, 52, 2013
|
|
4LD2
 
 | | Crystal structure of NE0047 in complex with cytidine | | 分子名称: | 1,2-ETHANEDIOL, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, Cytidine and deoxycytidylate deaminase zinc-binding region, ... | | 著者 | Bitra, A, Biswas, A, Anand, R. | | 登録日 | 2013-06-24 | | 公開日 | 2014-01-22 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural basis of the substrate specificity of cytidine deaminase superfamily Guanine deaminase
Biochemistry, 52, 2013
|
|
8IBO
 
 | |
8IBP
 
 | |
