4V4Y
 
 | | Crystal structure of the 70S Thermus thermophilus ribosome with translocated and rotated Shine-Dalgarno Duplex. | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Jenner, L, Yusupova, G, Rees, B, Moras, D, Yusupov, M. | | 登録日 | 2006-06-27 | | 公開日 | 2014-07-09 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (5.5 Å) | | 主引用文献 | Structural basis for messenger RNA movement on the ribosome.
Nature, 444, 2006
|
|
7CPZ
 
 | | Crystal structure of Streptoavidin-C1 from Streptomyces cinamonensis | | 分子名称: | BIOTIN, Mature Streptoavidin-C1 | | 著者 | Jeon, B.J, Kim, S, Lee, J.-H, Kim, M.S, Hwang, K.Y. | | 登録日 | 2020-08-08 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Iucrj, 8, 2021
|
|
7CQ0
 
 | | Crystal structure of Streptoavidin-C1 from Streptomyces cinamonensis | | 分子名称: | Mature Streptoavidin-C1 | | 著者 | Jeon, B.J, Kim, S, Lee, J.-H, Kim, M.S, Hwang, K.Y. | | 登録日 | 2020-08-08 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Insights into the structure of mature streptavidin C1 from Streptomyces cinnamonensis reveal the self-binding of the extension C-terminal peptide to biotin-binding sites.
Iucrj, 8, 2021
|
|
7CRN
 
 | |
7CUQ
 
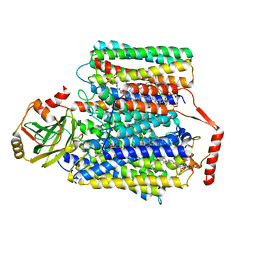 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | 著者 | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | 登録日 | 2020-08-24 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUW
 
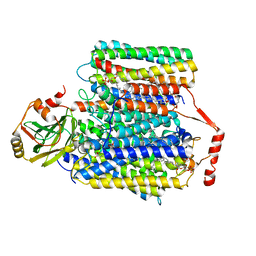 | | Ubiquinol Binding Site of Cytochrome bo3 from Escherichia coli | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | 著者 | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | 登録日 | 2020-08-25 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUB
 
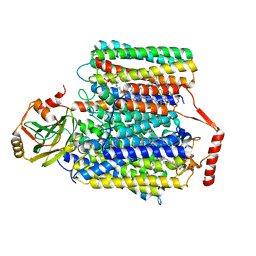 | | 2.55-Angstrom Cryo-EM structure of Cytochrome bo3 from Escherichia coli in Native Membrane | | 分子名称: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | 著者 | Li, J, Han, L, Gennis, R.B, Zhu, J.P, Zhang, K. | | 登録日 | 2020-08-22 | | 公開日 | 2021-08-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.55 Å) | | 主引用文献 | Cryo-EM structures of Escherichia coli cytochrome bo3 reveal bound phospholipids and ubiquinone-8 in a dynamic substrate binding site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CTM
 
 | |
7CWG
 
 | | Crystal structure of PDE8A catalytic domain in complex with 3a | | 分子名称: | 9-[(1~{S})-1-[4-[2,2-bis(fluoranyl)ethoxy]pyridin-2-yl]ethyl]-2-chloranyl-purin-6-amine, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | 著者 | Huang, Y, Wu, X.-N, Zhou, Q, Wu, Y, Luo, H.-B. | | 登録日 | 2020-08-28 | | 公開日 | 2021-09-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Rational Design of 2-Chloroadenine Derivatives as Highly Selective Phosphodiesterase 8A Inhibitors.
J.Med.Chem., 63, 2020
|
|
7CWF
 
 | | Crystal structure of PDE8A catalytic domain in complex with 2c | | 分子名称: | 9-[[4-[2,2-bis(fluoranyl)ethoxy]pyridin-2-yl]methyl]-2-chloranyl-purin-6-amine, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | 著者 | Huang, Y, Wu, X.-N, Zhou, Q, Wu, Y, Luo, H.-B. | | 登録日 | 2020-08-28 | | 公開日 | 2021-09-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Rational Design of 2-Chloroadenine Derivatives as Highly Selective Phosphodiesterase 8A Inhibitors.
J.Med.Chem., 63, 2020
|
|
8XGB
 
 | |
9FSV
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with epoxyketone inhibitor 42 | | 分子名称: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | 登録日 | 2024-06-22 | | 公開日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
9BRS
 
 | | Intact V-ATPase State 2 in synaptophysin knock-out isolated synaptic vesicles | | 分子名称: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | 著者 | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | 登録日 | 2024-05-11 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRU
 
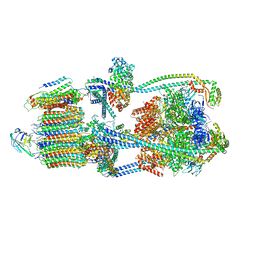 | | Intact V-ATPase State 1 in synaptophysin knock-out isolated synaptic vesicles | | 分子名称: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | 著者 | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | 登録日 | 2024-05-11 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BKK
 
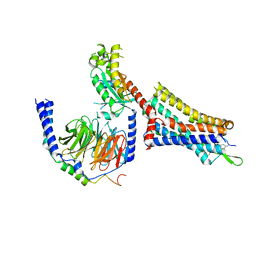 | | Cholecystokinin 1 receptor (CCK1R) sterol 7M mutant, Gq chimera (mGsqi) complex | | 分子名称: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Harikumar, K.G, Zhao, P, Cary, B.P, Xu, X, Desai, A.J, Mobbs, J.I, Toufaily, C, Furness, S.G.B, Christopoulos, A, Belousoff, M.J, Wootten, D, Sexton, P.M, Miller, L.J. | | 登録日 | 2024-04-29 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Cholesterol-dependent dynamic changes in the conformation of the type 1 cholecystokinin receptor affect ligand binding and G protein coupling.
Plos Biol., 22, 2024
|
|
9FST
 
 | | Yeast 20S proteasome with human beta1i (1-51) in complex with epoxyketone inhibitor LU-001i | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | 登録日 | 2024-06-21 | | 公開日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
To Be Published
|
|
9BKJ
 
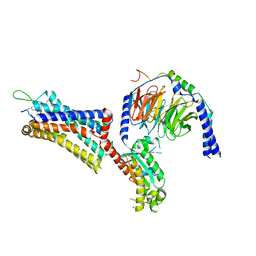 | | Cholecystokinin 1 receptor (CCK1R) Y140A mutant, Gq chimera (mGsqi) complex | | 分子名称: | AMINO GROUP, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | 著者 | Cary, B.P, Harikumar, K.G, Zhao, P, Desai, A.J, Mobbs, J.M, Toufaily, C, Furness, S.G.B, Christopoulos, A, Belousoff, M.J, Wootten, D, Sexton, P.M, Miller, L.J. | | 登録日 | 2024-04-29 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | Cholesterol-dependent dynamic changes in the conformation of the type 1 cholecystokinin receptor affect ligand binding and G protein coupling.
Plos Biol., 22, 2024
|
|
8ZC2
 
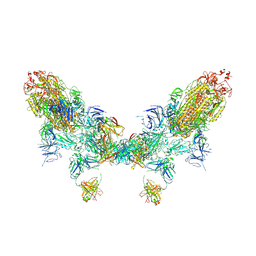 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with D1F6 Fab, head-to-head aggregate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | 著者 | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | 登録日 | 2024-04-28 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (7.82 Å) | | 主引用文献 | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
9FT1
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 13 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | 著者 | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | 登録日 | 2024-06-23 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
9FT0
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 42 | | 分子名称: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | 登録日 | 2024-06-23 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
6QKQ
 
 | |
6QKP
 
 | |
9BRQ
 
 | | Intact V-ATPase State 3 and synaptophysin complex in mouse brain isolated synaptic vesicles | | 分子名称: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | 著者 | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | 登録日 | 2024-05-11 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
6Q44
 
 | | Est3 telomerase subunit in the yeast Hansenula polymorpha | | 分子名称: | Uncharacterized protein | | 著者 | Mantsyzov, A.B, Mariasina, S.S, Petrova, O.A, Efimov, S.V, Dontsova, O.A, Polshakov, V.I. | | 登録日 | 2018-12-05 | | 公開日 | 2019-12-25 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Insights into the structure and function of Est3 from the Hansenula polymorpha telomerase.
Sci Rep, 10, 2020
|
|
9BRR
 
 | | Intact V-ATPase State 3 in synaptophysin knock-out isolated synaptic vesicles | | 分子名称: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | 著者 | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | 登録日 | 2024-05-11 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
