8XAU
 
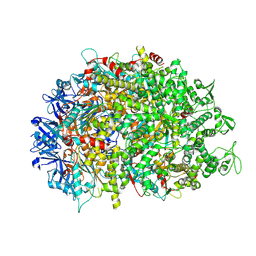 | | Cryo-EM structure of HerA | | 分子名称: | ATP-binding protein | | 著者 | Wang, Y, Deng, Z. | | 登録日 | 2023-12-05 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 2024
|
|
7SC5
 
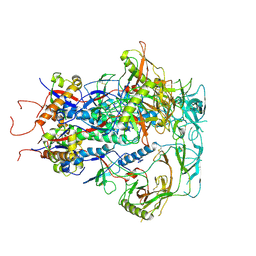 | | Cytoplasmic tail deleted HIV Env trimer in nanodisc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | 著者 | Yang, S, Walz, T. | | 登録日 | 2021-09-27 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | Dynamic HIV-1 spike motion creates vulnerability for its membrane-bound tripod to antibody attack.
Nat Commun, 13, 2022
|
|
7SD3
 
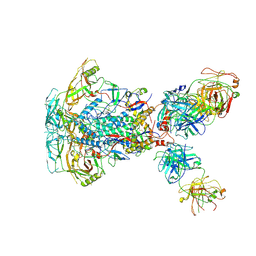 | | Cytoplasmic tail deleted HIV-1 Env bound with three 4E10 Fabs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4E10 Fab heavy chain, ... | | 著者 | Yang, S, Walz, T. | | 登録日 | 2021-09-29 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Dynamic HIV-1 spike motion creates vulnerability for its membrane-bound tripod to antibody attack.
Nat Commun, 13, 2022
|
|
8XAV
 
 | |
8XAW
 
 | |
8VGR
 
 | |
8VJS
 
 | |
8VJR
 
 | |
7SIS
 
 | |
8IOI
 
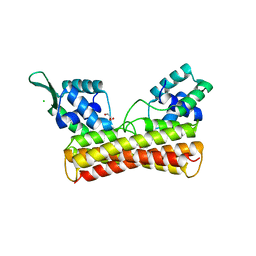 | | Crystal Structure of PadR- family transcriptional regulator Rv1176c from Mycobacterium tuberculosis H37Rv. | | 分子名称: | CHLORIDE ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | 著者 | Yadav, V, Zohib, M, Pal, R.K, Biswal, B.K, Arora, A. | | 登録日 | 2023-03-11 | | 公開日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Structural and biophysical characterization of PadR family protein Rv1176c of Mycobacterium tuberculosis H37Rv.
Int.J.Biol.Macromol., 263, 2024
|
|
8XAY
 
 | |
1I6Q
 
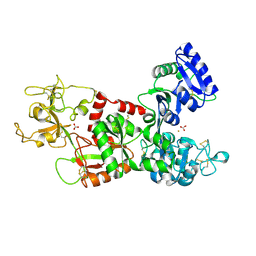 | | Formation of a protein intermediate and its trapping by the simultaneous crystallization process: Crystal structure of an iron-saturated intermediate in the FE3+ binding pathway of camel lactoferrin at 2.7 resolution | | 分子名称: | CARBONATE ION, FE (III) ION, LACTOFERRIN | | 著者 | Khan, J.A, Kumar, P, Srinivasan, A, Singh, T.P. | | 登録日 | 2001-03-03 | | 公開日 | 2001-11-07 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Protein intermediate trapped by the simultaneous crystallization process. Crystal structure of an iron-saturated intermediate in the Fe3+ binding pathway of camel lactoferrin at 2.7 a resolution.
J.Biol.Chem., 276, 2001
|
|
7S3V
 
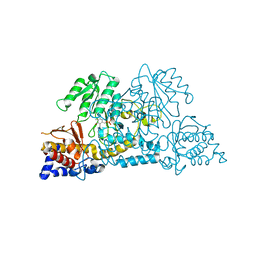 | |
6VEP
 
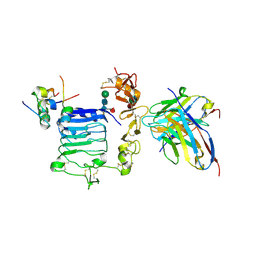 | |
7SOD
 
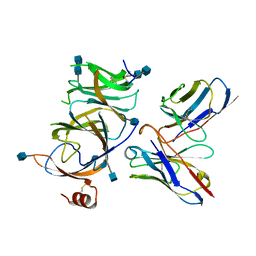 | |
7SOB
 
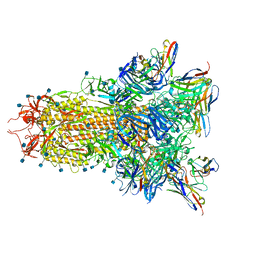 | |
7SO9
 
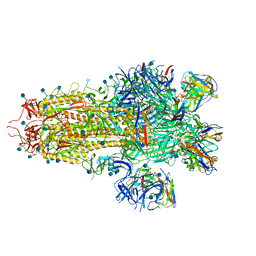 | |
7SOC
 
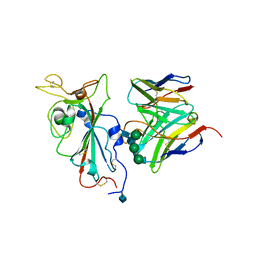 | |
7SOE
 
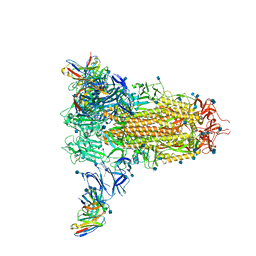 | |
1ELP
 
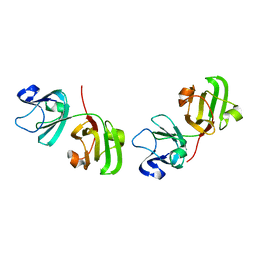 | | GAMMA-D CRYSTALLIN STRUCTURE AT 1.95 A RESOLUTION | | 分子名称: | GAMMA-D CRYSTALLIN | | 著者 | Chirgadze, Yu.N, Driessen, H.P.C, Wright, G, Slingsby, C, Hay, R.E, Lindley, P.F. | | 登録日 | 1995-12-20 | | 公開日 | 1996-06-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure of bovine eye lens gammaD (gammaIIIb)-crystallin at 1.95 A.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
7SOF
 
 | |
7SOA
 
 | |
7SZ1
 
 | | Cryo-EM structure of the extracellular module of the full-length EGFR L834R bound to EGF. "tips-separated" conformation | | 分子名称: | Epidermal growth factor, Epidermal growth factor receptor | | 著者 | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | 登録日 | 2021-11-25 | | 公開日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SYD
 
 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to EGF "tips-juxtaposed" conformation | | 分子名称: | Epidermal growth factor, Epidermal growth factor receptor | | 著者 | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | 登録日 | 2021-11-24 | | 公開日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
7SYE
 
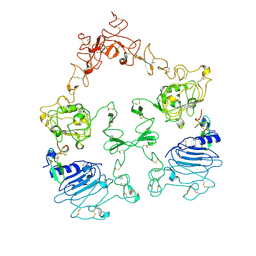 | | Cryo-EM structure of the extracellular module of the full-length EGFR bound to EGF. "tips-separated" conformation | | 分子名称: | Epidermal growth factor, Epidermal growth factor receptor | | 著者 | Huang, Y, Ognjenovic, J, Karandur, D, Miller, K, Merk, A, Subramaniam, S, Kuriyan, J. | | 登録日 | 2021-11-24 | | 公開日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor.
Elife, 10, 2021
|
|
